全新设计的蛋白质能中和致命的蛇毒毒素
IF 48.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
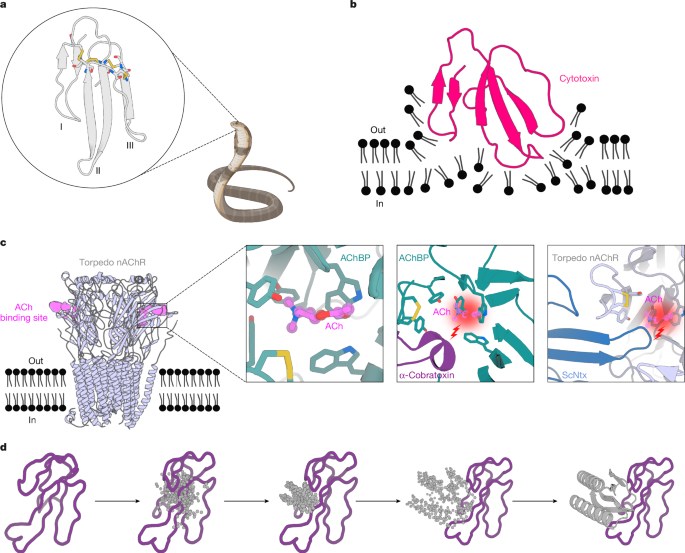
蛇咬伤仍然是一种毁灭性的、被忽视的热带疾病,每年夺去超过10万人的生命,并造成严重的并发症和更多人的长期残疾。三指毒素(Three-finger toxin, 3FTx)是一种高毒性成分,可引起多种病理,包括严重的组织损伤和尼古丁乙酰胆碱受体的抑制,从而导致危及生命的神经毒性。目前,治疗蛇咬伤的唯一方法是从免疫动物的血浆中提取多克隆抗体,这些抗体对3ftxs5,6,7的治疗成本高且效果有限。在这里,我们使用深度学习方法重新设计蛋白来结合来自3FTx家族的短链和长链α-神经毒素和细胞毒素。通过有限的实验筛选,我们获得了具有显著热稳定性、高结合亲和力和与计算模型接近原子水平一致性的蛋白质设计。设计的蛋白在体外有效地中和了所有三个3FTx亚家族,并保护小鼠免受致命的神经毒素攻击。这种有效、稳定且易于制造的毒素中和蛋白可以为更安全、成本效益高且易于获得的下一代抗蛇毒血清治疗提供基础。除了蛇咬伤之外,我们的研究结果强调了计算设计如何通过大幅降低被忽视的热带病治疗方法开发的成本和资源需求,帮助实现治疗发现的民主化,特别是在资源有限的情况下。本文章由计算机程序翻译,如有差异,请以英文原文为准。

De novo designed proteins neutralize lethal snake venom toxins
Snakebite envenoming remains a devastating and neglected tropical disease, claiming over 100,000 lives annually and causing severe complications and long-lasting disabilities for many more1,2. Three-finger toxins (3FTx) are highly toxic components of elapid snake venoms that can cause diverse pathologies, including severe tissue damage3 and inhibition of nicotinic acetylcholine receptors, resulting in life-threatening neurotoxicity4. At present, the only available treatments for snakebites consist of polyclonal antibodies derived from the plasma of immunized animals, which have high cost and limited efficacy against 3FTxs5–7. Here we used deep learning methods to de novo design proteins to bind short-chain and long-chain α-neurotoxins and cytotoxins from the 3FTx family. With limited experimental screening, we obtained protein designs with remarkable thermal stability, high binding affinity and near-atomic-level agreement with the computational models. The designed proteins effectively neutralized all three 3FTx subfamilies in vitro and protected mice from a lethal neurotoxin challenge. Such potent, stable and readily manufacturable toxin-neutralizing proteins could provide the basis for safer, cost-effective and widely accessible next-generation antivenom therapeutics. Beyond snakebite, our results highlight how computational design could help democratize therapeutic discovery, particularly in resource-limited settings, by substantially reducing costs and resource requirements for the development of therapies for neglected tropical diseases. Deep learning methods have been used to design proteins that can neutralize the effects of three-finger toxins found in snake venom, which could lead to the development of safer and more accessible antivenom treatments.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


