全球人群肠道微生物群中肠杆菌科的生态动态
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
来自肠杆菌科的肠道细菌是世界范围内机会性感染的主要原因。鉴于它们在健康人类肠道微生物群中的普遍存在,种间相互作用可能在调节感染抗性中发挥作用。在这里,我们通过利用跨越45个国家的12238个公共人类肠道宏基因组的大规模数据集,揭示了与肠杆菌科定植和丰度相关的全球生态模式。机器学习分析确定了与肠杆菌科定植状态相关的强大肠道微生物组特征,该特征在健康状态和地理位置上是一致的。我们将172种肠道微生物分类为共定植菌,135种为共排斥菌,揭示了Faecalimonas phoceensis在全属范围内的定植抗性信号和菌株特异性的共定植模式。共排斥与短链脂肪酸产生、铁代谢和群体感应的功能有关,而共定植与肠杆菌科更大的功能多样性和代谢相似性有关。我们的工作强调了肠道环境在肠道相关机会性病原体定植成功中的关键作用,这对开发非抗生素治疗策略具有重要意义。本文章由计算机程序翻译,如有差异,请以英文原文为准。

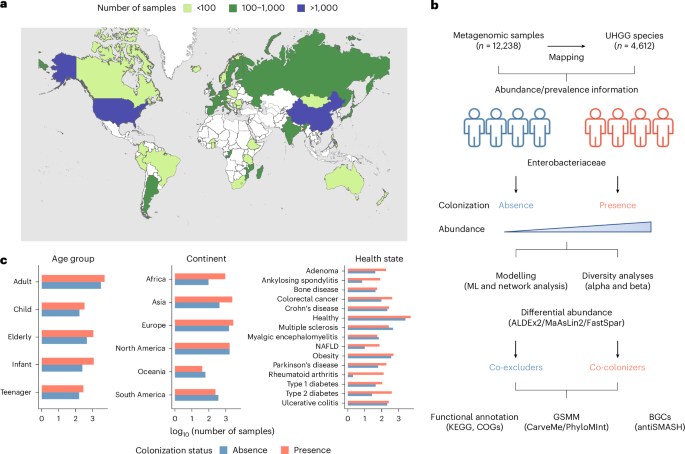
Ecological dynamics of Enterobacteriaceae in the human gut microbiome across global populations
Gut bacteria from the Enterobacteriaceae family are a major cause of opportunistic infections worldwide. Given their prevalence among healthy human gut microbiomes, interspecies interactions may play a role in modulating infection resistance. Here we uncover global ecological patterns linked to Enterobacteriaceae colonization and abundance by leveraging a large-scale dataset of 12,238 public human gut metagenomes spanning 45 countries. Machine learning analyses identified a robust gut microbiome signature associated with Enterobacteriaceae colonization status, consistent across health states and geographic locations. We classified 172 gut microbial species as co-colonizers and 135 as co-excluders, revealing a genus-wide signal of colonization resistance within Faecalibacterium and strain-specific co-colonization patterns of the underexplored Faecalimonas phoceensis. Co-exclusion is linked to functions involved in short-chain fatty acid production, iron metabolism and quorum sensing, while co-colonization is linked to greater functional diversity and metabolic resemblance to Enterobacteriaceae. Our work underscores the critical role of the intestinal environment in the colonization success of gut-associated opportunistic pathogens with implications for developing non-antibiotic therapeutic strategies. Assessing more than 12,000 metagenomic samples from across the world using computational approaches, the authors determined interactions between species that co-colonize or co-exclude Enterobacteriaceae in terms of functional pathways and metabolites in healthy human gut microbiomes.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


