使用长读测序分析表观基因组
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
牛津纳米孔技术公司和太平洋生物科学公司的单分子长读测序(LRS)技术的出现彻底改变了基因组学、转录组学以及最近的表观基因组学研究。这些技术具有明显的优势,包括直接检测甲基化DNA和同时评估跨越多个千碱基的DNA序列以及它们在单分子水平上的修饰。这使得分析染色质状态的新方法得以发展,并使整合DNA甲基化、染色质可及性、转录因子结合和组蛋白修饰的数据成为可能,从而促进全面的表观基因组分析。由于最近的进展,可以使用LRS方法检测替代的,新生的和翻译的转录本。本综述讨论了基于lrs的表征染色质状态的实验和计算策略,并强调了它们相对于短读测序方法的优势。此外,我们展示了如何将各种长读方法集成到设计多组学研究中,以研究染色质状态和转录动力学之间的关系。本文章由计算机程序翻译,如有差异,请以英文原文为准。
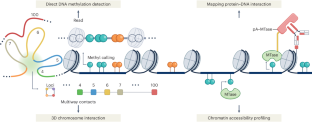

Profiling the epigenome using long-read sequencing
The advent of single-molecule, long-read sequencing (LRS) technologies by Oxford Nanopore Technologies and Pacific Biosciences has revolutionized genomics, transcriptomics and, more recently, epigenomics research. These technologies offer distinct advantages, including the direct detection of methylated DNA and simultaneous assessment of DNA sequences spanning multiple kilobases along with their modifications at the single-molecule level. This has enabled the development of new assays for analyzing chromatin states and made it possible to integrate data for DNA methylation, chromatin accessibility, transcription factor binding and histone modifications, thereby facilitating comprehensive epigenomic profiling. Owing to recent advancements, alternative, nascent and translating transcripts can be detected using LRS approaches. This Review discusses LRS-based experimental and computational strategies for characterizing chromatin states and highlights their advantages over short-read sequencing methods. Furthermore, we demonstrate how various long-read methods can be integrated to design multi-omics studies to investigate the relationship between chromatin states and transcriptional dynamics. Long-read sequencing technologies have revolutionized genomics and transcriptomics, and more recently enabled comprehensive epigenomic profiling. These advances now also allow investigation of the relationship between chromatin states and transcriptional dynamics.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


