一个寿命可推广的头骨剥离模型磁共振图像,利用大脑地图集的先验知识
IF 26.8
1区 医学
Q1 ENGINEERING, BIOMEDICAL
引用次数: 0
摘要
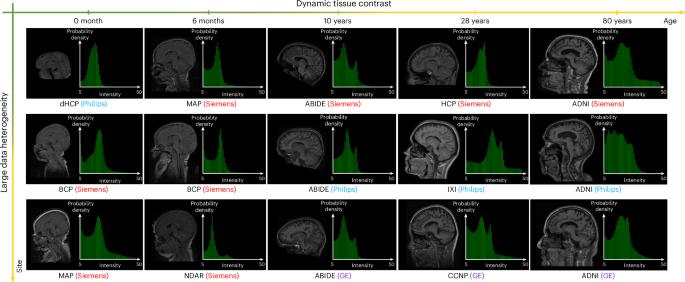
在大脑的磁共振成像中,成像预处理步骤从图像中去除头骨和其他非脑组织。但是,这种头骨剥离过程的方法经常与医疗站点之间的大量数据异质性以及寿命期间组织对比的动态变化作斗争。在这里,我们报告了一个磁共振图像的头骨剥离模型,该模型通过利用来自大脑图谱的个性化先验来概括整个生命周期。该模型由一个大脑提取模块和一个注册模块组成,前者提供图像上脑组织的初始估计,后者从特定年龄的图谱中获得个性化先验。该模型比最先进的颅骨剥离方法要准确得多,正如我们从18个地点通过各种成像协议和扫描仪获得的21,334个寿命的大型多样化数据集所展示的那样,它产生了自然一致和无缝的脑容量寿命变化,忠实地描绘了大脑发育和衰老的潜在生物过程。本文章由计算机程序翻译,如有差异,请以英文原文为准。
A lifespan-generalizable skull-stripping model for magnetic resonance images that leverages prior knowledge from brain atlases
In magnetic resonance imaging of the brain, an imaging-preprocessing step removes the skull and other non-brain tissue from the images. But methods for such a skull-stripping process often struggle with large data heterogeneity across medical sites and with dynamic changes in tissue contrast across lifespans. Here we report a skull-stripping model for magnetic resonance images that generalizes across lifespans by leveraging personalized priors from brain atlases. The model consists of a brain extraction module that provides an initial estimation of the brain tissue on an image, and a registration module that derives a personalized prior from an age-specific atlas. The model is substantially more accurate than state-of-the-art skull-stripping methods, as we show with a large and diverse dataset of 21,334 lifespans acquired from 18 sites with various imaging protocols and scanners, and it generates naturally consistent and seamless lifespan changes in brain volume, faithfully charting the underlying biological processes of brain development and ageing. A skull-stripping model for magnetic resonance images that leverages personalized priors from atlases of brain scans generates naturally consistent and seamless lifespan changes in brain volume.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Biomedical Engineering
Medicine-Medicine (miscellaneous)
CiteScore
45.30
自引率
1.10%
发文量
138
期刊介绍:
Nature Biomedical Engineering is an online-only monthly journal that was launched in January 2017. It aims to publish original research, reviews, and commentary focusing on applied biomedicine and health technology. The journal targets a diverse audience, including life scientists who are involved in developing experimental or computational systems and methods to enhance our understanding of human physiology. It also covers biomedical researchers and engineers who are engaged in designing or optimizing therapies, assays, devices, or procedures for diagnosing or treating diseases. Additionally, clinicians, who make use of research outputs to evaluate patient health or administer therapy in various clinical settings and healthcare contexts, are also part of the target audience.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


