端粒到端粒羊基因组组装鉴定与羊毛细度相关的变异
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
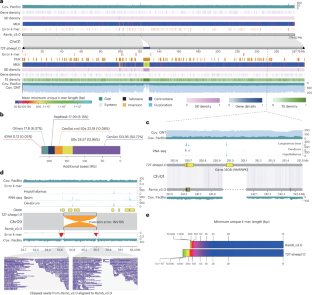
正在进行的改进绵羊参考基因组组装的努力仍然留下许多空白和不完整的区域,导致基因组研究中一些常见的失败和错误。在这里,我们报告了一只公羊(T2T-sheep1.0)的2.85 gb无间隙端粒到端粒基因组,包括所有常染色体和X染色体和Y染色体。该基因组在最新的参考序列ARS-UI_Ramb_v3.0上增加了220.05 Mb先前未解析的区域和754个新基因;在着丝区包含四种类型的重复单元(SatI、SatII、SatIII和CenY)。T2T-sheep1.0的碱基精度超过99.999%,修正了以前参考序列中的一些结构错误,提高了重复序列的结构变异检测。将全球家羊和野生羊的全基因组短序列与T2T-sheep1.0进行比对,在以前未解决的区域发现了2664979个新的单核苷酸多态性,这改善了群体遗传分析和驯化选择性信号(如ABCC4)和羊毛细度(如FOXQ1)的检测。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Telomere-to-telomere sheep genome assembly identifies variants associated with wool fineness
Ongoing efforts to improve sheep reference genome assemblies still leave many gaps and incomplete regions, resulting in a few common failures and errors in genomic studies. Here, we report a 2.85-Gb gap-free telomere-to-telomere genome of a ram (T2T-sheep1.0), including all autosomes and the X and Y chromosomes. This genome adds 220.05 Mb of previously unresolved regions and 754 new genes to the most updated reference assembly ARS-UI_Ramb_v3.0; it contains four types of repeat units (SatI, SatII, SatIII and CenY) in centromeric regions. T2T-sheep1.0 has a base accuracy of more than 99.999%, corrects several structural errors in previous reference assemblies and improves structural variant detection in repetitive sequences. Alignment of whole-genome short-read sequences of global domestic and wild sheep against T2T-sheep1.0 identifies 2,664,979 new single-nucleotide polymorphisms in previously unresolved regions, which improves the population genetic analyses and detection of selective signals for domestication (for example, ABCC4) and wool fineness (for example, FOXQ1). A gap-free telomere-to-telomere genome assembly for a ram of Hu sheep uncovers genes and variants associated with domestication and selection for wool fineness.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


