小麦660 K和90 K SNP序列的调和及其在面包小麦面团流变学特性中的应用
IF 11.4
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
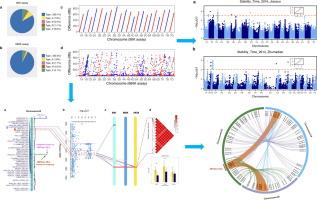
高密度小麦660 K和90 K SNP阵列是了解小麦性状遗传基础的有力工具。然而,由于不同版本的中国春季基因组在构建阵列过程中导致了它们的物理位置不一致,这给进一步的应用带来了混乱和不便。目的随着小麦基因组测序的快速发展,对小麦660 K和90 K SNP序列进行比对,揭示面包小麦面团流变学特性的遗传基础。方法在最近发布的小麦品种AK58基因组中,对小麦660 K和90 K SNP阵列进行物理定位。接下来,我们分别使用更新和未更新的阵列进行了全基因组关联研究(GWAS)和连锁分析,以确定与质量性状相关的重要遗传位点。结果精化结果显示,小麦660 K和90 K SNP阵列中分别有92.3 %和83 %的SNP被精确定位到AK58基因组。更新后的660个 K和90个 K阵列的GWAS结果表明,在多个环境中,由1032个显著snp组成的26个区间与9个品质性状相关。使用更新的阵列时,1D上稳定时间的显著间隔缩小到8.4 Mb,而使用未更新的阵列时,该间隔为405 Mb。连锁分析发现一个重要的QTL QST.henau-1D。2稳定时间为1.64 Mb。通过共线性分析,GWAS和QTL的整合结果将显著区间缩小至6.46 Mb,共包含35个注释基因。经过t检验和基因表达分析,其中7个是潜在的候选基因,从而确定了有利的单倍型,有利于标记辅助选择。结论小麦660 K和90 K阵列的调合促进了它们的高效应用。本研究发现的重要遗传位点和有利单倍型为小麦品质育种提供了有价值的信息。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Reconciliation of wheat 660 K and 90 K SNP arrays and their utilization in dough rheological properties of bread wheat
Introduction
High-density Wheat 660 K and 90 K SNP arrays are powerful tools for understanding the genetic basis of wheat traits. However, their inconsistantly physical positions that were caused by different versions of Chinese Spring genome during developing arrays are confused and inconvenient for further application.Objective
With the repid development of wheat geonome sequencing, we aim to reconciliate Wheat 660 K and 90 K SNP arrays in modern cultivar and reveal the genetic basis of dough rheological properties in bread wheat.Methods
We refined physical positions of Wheat 660 K and 90 K SNP arrays in the currently popular wheat cultivar AK58 genome that was released more recently. We next performed genome-wide association studies (GWAS) and linkage analysis to identify important genetic loci related to quality traits using updated and un-updated arrays, respectively.Results
Refining results showed that 92.3 % and 83 % of SNPs in the Wheat 660 K and 90 K SNP arrays were precisely mapped to the AK58 genome, respective. GWAS results by the updated 660 K and 90 K arrays indicated that 26 intervals composed of 1032 significant SNPs were associated with 9 quality traits in multiple environments. The significant interval for stability time on 1D was narrowed into an 8.4-Mb region using the updated arrays, whereas the interval is 405 Mb using the un-updated arrays. Linkage analysis revealed an important QTL QST.henau-1D.2 for stability time with 1.64 Mb. Integration of GWAS and QTL results narrowed the significant interval into 6.46 Mb containing 35 annotation genes by collinearity analysis. After T-test, gene expression analysis, seven of them are potential candidate genes and thus favorable haplotypes are identified to benefit marker-assisted selection.Conclusion
A reconciliation of Wheat 660 K and 90 K arrays promote their efficient applications. Important genetic loci and favorable haplotypes identified in this study provided valuable information for wheat quality breeding.求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Advanced Research
Multidisciplinary-Multidisciplinary
CiteScore
21.60
自引率
0.90%
发文量
280
审稿时长
12 weeks
期刊介绍:
Journal of Advanced Research (J. Adv. Res.) is an applied/natural sciences, peer-reviewed journal that focuses on interdisciplinary research. The journal aims to contribute to applied research and knowledge worldwide through the publication of original and high-quality research articles in the fields of Medicine, Pharmaceutical Sciences, Dentistry, Physical Therapy, Veterinary Medicine, and Basic and Biological Sciences.
The following abstracting and indexing services cover the Journal of Advanced Research: PubMed/Medline, Essential Science Indicators, Web of Science, Scopus, PubMed Central, PubMed, Science Citation Index Expanded, Directory of Open Access Journals (DOAJ), and INSPEC.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


