tRNA基因表达的适应性缺失导致海洋蓝聚球菌的噬菌体抗性
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
聚珠球菌是海洋中重要的初级生产者,与噬藻体共存,后者是重要的致死率。细菌对噬菌体感染的耐药性是一个非常有趣的话题,但对生态相关系统知之甚少。本研究利用外源基因表达和基因破坏研究海洋聚球菌WH5701对Syn9噬藻体的细胞内抗性机制。聚球菌WH5701所具有的限制性修饰和Gabija防御系统对耐药无促进作用。相反,耐药主要是由LeuTAA tRNA水平不足驱动的,在被动的细胞内耐药模式下阻止了关键噬菌体基因的翻译。恢复细胞tRNA表达使蓝藻对感染敏感。我们提出了一种进化情景,即细胞密码子使用的变化、噬菌体获得tRNA以及细胞和噬菌体tRNA表达的丧失导致了一种有效的耐药性手段,强调了细菌和噬菌体在形成其共同进化轨迹方面的动态相互作用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
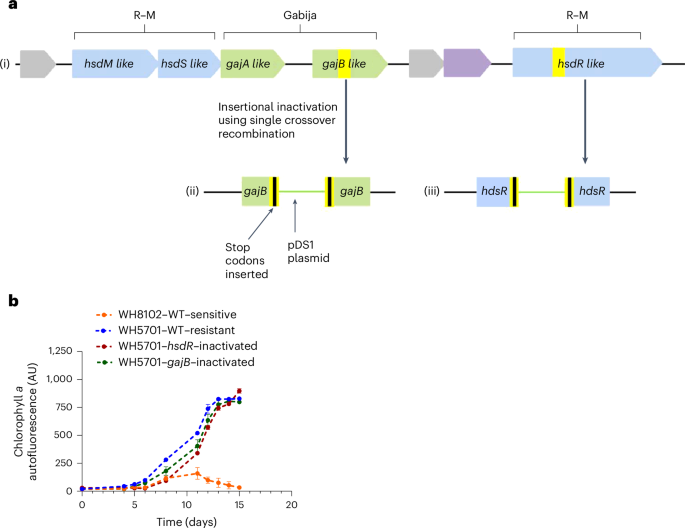

Adaptive loss of tRNA gene expression leads to phage resistance in a marine Synechococcus cyanobacterium
Synechococcus is a significant primary producer in the oceans, coexisting with cyanophages, which are important agents of mortality. Bacterial resistance against phage infection is a topic of significant interest, yet little is known for ecologically relevant systems. Here we use exogenous gene expression and gene disruption to investigate mechanisms underlying intracellular resistance of marine Synechococcus WH5701 to the Syn9 cyanophage. The restriction–modification and Gabija defence systems possessed by Synechococcus WH5701 did not contribute to resistance. Instead, resistance was primarily driven by insufficient levels of LeuTAA tRNA, preventing translation of key phage genes in a passive, intracellular mode of resistance. Restoring cellular tRNA expression rendered the cyanobacterium sensitive to infection. We propose an evolutionary scenario whereby changes in cell codon usage, acquisition of tRNAs by the phage and loss of cell and phage tRNA expression resulted in an effective means of resistance, highlighting the dynamic interplay between bacteria and phages in shaping their co-evolutionary trajectories. Depletion of host LeuTAA tRNA levels prevents the translation of key cyanophage genes during infection and represents a passive, intracellular mode of resistance with implications for co-evolution.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


