细菌IS110转座子中古细菌和真核生物RNA引导RNA修饰的进化起源
IF 19.4
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
转座酶基因普遍存在于生命的各个领域,并为新蛋白质功能的进化提供了丰富的储备。在这里,我们报告了细菌is110家族转座酶和古细菌/真核生物nop5家族蛋白之间的深层进化联系,前者通过桥接RNA催化RNA引导的DNA重组,后者通过C/D-box snoRNAs促进RNA引导的RNA 2 ' - o -甲基化。基于蛋白质序列的保守性、结构域结构、三维结构和非编码RNA的特征,以及系统发育分析,我们提出可编程RNA修饰是通过提取来自is110样转座子的成分而出现的。这些发现强调了转座因子的反复驯化事件如何推动rna引导机制的进化。本文章由计算机程序翻译,如有差异,请以英文原文为准。
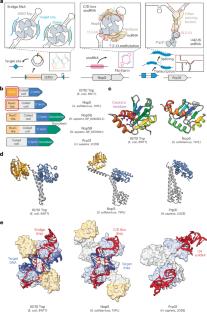

Evolutionary origins of archaeal and eukaryotic RNA-guided RNA modification in bacterial IS110 transposons
Transposase genes are ubiquitous in all domains of life and provide a rich reservoir for the evolution of novel protein functions. Here we report deep evolutionary links between bacterial IS110-family transposases, which catalyse RNA-guided DNA recombination using bridge RNAs, and archaeal/eukaryotic Nop5-family proteins, which promote RNA-guided RNA 2′-O-methylation using C/D-box snoRNAs. On the basis of conservation of protein sequence, domain architecture, three-dimensional structure and non-coding RNA features, alongside phylogenetic analyses, we propose that programmable RNA modification emerged through the exaptation of components derived from IS110-like transposons. These findings underscore how recurrent domestication events of transposable elements have driven the evolution of RNA-guided mechanisms. Structural and phylogenetic analyses show that programmable RNA modifications ubiquitous in archaea and eukarya arose from bacterial transposons.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


