对西伯利亚蜱传脑炎病毒(TBEV)的全基因组测序监测在吉尔吉斯斯坦发现了一个额外的谱系。
IF 2.7
4区 医学
Q3 VIROLOGY
引用次数: 0
摘要
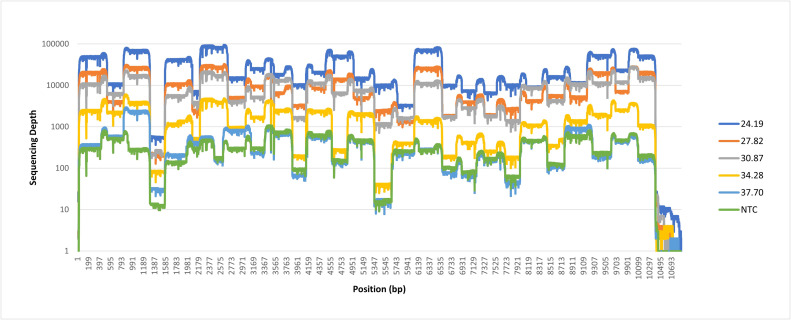
蜱传脑炎病毒(TBEV)是欧洲和亚洲最流行的蜱传病毒性疾病。该病毒有三种主要亚型:欧洲、西伯利亚和远东,每一种都有独特的生态、临床表现和地理分布。近年来,其他的TBEV亚型也被描述,即喜马拉雅亚型和贝加尔湖亚型。已经描述了乙型脑炎病毒亚型之间的毒力差异,其中远东亚型在人类中引起最严重的疾病。考虑到新的热带病疫源地的出现,在流行地区对病毒的遗传特征进行描述不仅对于更好地了解其流行病学,而且对于确定可能的毒力遗传决定因素以及开发准确的诊断和治疗方法至关重要。在我们之前的研究中,我们确定了吉尔吉斯共和国(吉尔吉斯斯坦)的六个地区的TBEV,以及Ala-Archa国家自然公园是TBEV传播的焦点。虽然我们能够从吉尔吉斯斯坦的过sulcatus蜱中检索到第一个部分TBEV序列,但当时我们无法检索到完整的基因组序列。在这项研究中,我们利用序列独立的单引物扩增(SISPA)方案,检索了我们之前2009年的TBEV蜱虫样本(菌株KY09)的完整基因组序列,产生了来自吉尔吉斯斯坦的第三个完整的TBEV基因组,以及Vasilchenko谱系中聚集区域的第一个基因组,表明该谱系的分布比之前认为的更广泛。我们还开发了一种西伯利亚TBEV (TBEV- sib)的平铺扩增子方案,该方案在100倍测序深度下,对低至1.13×104 RNA拷贝/ml的样品产生bbb90 %的参考覆盖率。由于高病毒载量在TBEV临床样本中是罕见的,开发的方案通过提供一套新的引物在测序之前进一步扩增病毒基因组,为TBEV- sib流行区域增加了价值。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Whole-genome sequencing surveillance of Siberian tick-borne encephalitis virus (TBEV) identifies an additional lineage in Kyrgyzstan
Tick-borne encephalitis virus (TBEV) is the most prevalent tick-borne viral disease in Europe and Asia. There are three main subtypes of the virus: European, Siberian, and Far Eastern, each of which having distinctive ecology, clinical presentation, and geographic distribution. In recent years, other TBEV subtypes have been described, namely the Himalayan and Baikalian subtypes. Differences in virulence between TBEV subtypes have been described, with the Far Eastern subtype causing the most severe disease in humans. Considering the emergence of new TBEV foci, the genetic characterisation of the virus in endemic regions is crucial to not only better understand its epidemiology, but also to identify possible genetic determinants of virulence, as well as develop accurate diagnostics and therapeutics.
In our previous study, we identified TBEV in six localities of the Kyrgyz Republic (Kyrgyzstan), and Ala-Archa National Nature Park as a focus of TBEV transmission. Whilst we were able to retrieve the first partial TBEV sequence from Kyrgyzstan from Ixodes persulcatus ticks, we were unable to retrieve a complete genome sequence at that time.
In this study, we have utilised a sequence-independent single-primer amplification (SISPA) protocol and retrieved the complete genome sequence of our previous 2009 TBEV tick sample (strain KY09) producing the third complete TBEV genome from Kyrgyzstan, and the first genome from the region clustering within the Vasilchenko lineage, suggesting a wider distribution for the lineage than was previously thought.
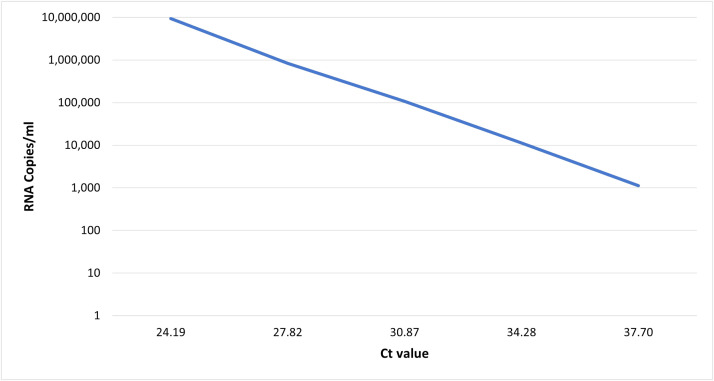
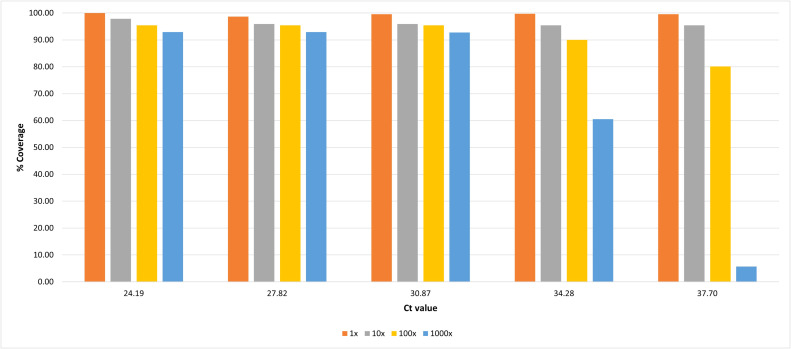
We have also developed a tiling amplicon scheme for Siberian TBEV (TBEV-Sib) which produced > 90 % reference coverage at 100x sequencing depths for samples with as little as 1.13×104 RNA copies/ml. Since high viral loads are rare in TBEV clinical samples, the developed protocol adds value to TBEV-Sib endemic regions by offering a novel set of primers to further amplify the viral genome prior to sequencing.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Virus research
医学-病毒学
CiteScore
9.50
自引率
2.00%
发文量
239
审稿时长
43 days
期刊介绍:
Virus Research provides a means of fast publication for original papers on fundamental research in virology. Contributions on new developments concerning virus structure, replication, pathogenesis and evolution are encouraged. These include reports describing virus morphology, the function and antigenic analysis of virus structural components, virus genome structure and expression, analysis on virus replication processes, virus evolution in connection with antiviral interventions, effects of viruses on their host cells, particularly on the immune system, and the pathogenesis of virus infections, including oncogene activation and transduction.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


