核小体纤维拓扑结构引导转录因子与增强子结合
IF 50.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
细胞特性需要多种转录因子(TFs)的协同作用,这些转录因子与细胞类型特异性基因的增强子结合在一起。尽管转录因子能识别可访问染色质中的特定 DNA 基团,但这一信息不足以解释转录因子如何选择增强子1。在这里,我们比较了诱导不同细胞状态的四种不同的TF组合,分析了TF基因组占有率、染色质可及性、核糖体定位和核糖体分辨率下的三维基因组组织。我们的研究表明,单核糖体上的主题识别只能破译 TF 的单独结合。当结合在一起时,TF 相互合作或竞争,以具有确定三维组织的核糖体阵列为目标,以特定模式显示主题。在一种组合中,图案的方向性将 TF 组合结合沿染色质环漏斗状排列,然后横向渗入相邻的增强子。在其他组合中,TF集结在主题密集且高度互联的环连接点上,随后转移到邻近的系特异性位点。我们提出了一种引导-搜索模型,在该模型中,核小体纤维上的主题语法就像路标元素一样,引导 TF 组合结合到增强子上。本文章由计算机程序翻译,如有差异,请以英文原文为准。

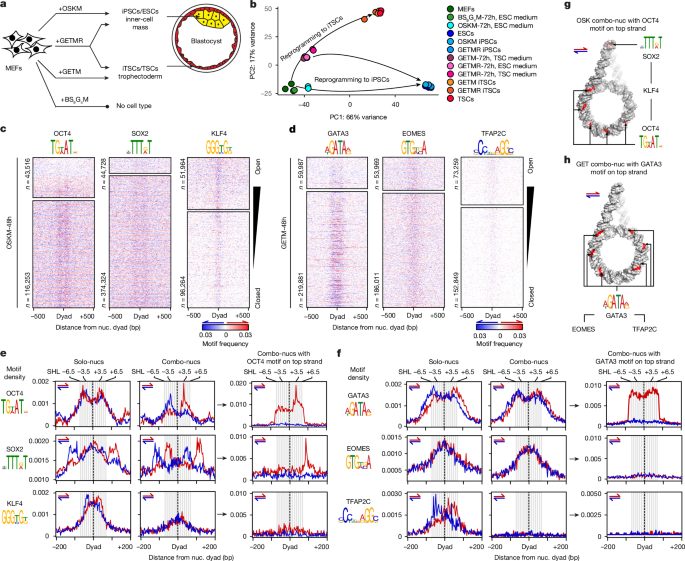
Nucleosome fibre topology guides transcription factor binding to enhancers
Cellular identity requires the concerted action of multiple transcription factors (TFs) bound together to enhancers of cell-type-specific genes. Despite TFs recognizing specific DNA motifs within accessible chromatin, this information is insufficient to explain how TFs select enhancers1. Here we compared four different TF combinations that induce different cell states, analysing TF genome occupancy, chromatin accessibility, nucleosome positioning and 3D genome organization at the nucleosome resolution. We show that motif recognition on mononucleosomes can decipher only the individual binding of TFs. When bound together, TFs act cooperatively or competitively to target nucleosome arrays with defined 3D organization, displaying motifs in particular patterns. In one combination, motif directionality funnels TF combinatorial binding along chromatin loops, before infiltrating laterally to adjacent enhancers. In other combinations, TFs assemble on motif-dense and highly interconnected loop junctions, and subsequently translocate to nearby lineage-specific sites. We propose a guided-search model in which motif grammar on nucleosome fibres acts as signpost elements, directing TF combinatorial binding to enhancers. Motif grammar on nucleosome fibres acts as signpost elements, directing TF combinatorial binding to enhancers.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


