伪对称蛋白质纳米笼的分层设计
IF 48.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
大小从数百千道尔顿到数百兆道尔顿不等的离散蛋白质组装体是生物系统中无处不在的特征,并能执行高度专业化的功能1,2。尽管最近在精确设计新的自组装蛋白质方面取得了令人瞩目的进展,但这些组装体的大小和复杂性一直受到严格对称性的限制3。在此,我们从细菌微腔和病毒外壳中观察到的假对称性得到启发,开发出一种分层计算方法,用于设计大型假对称自组装蛋白质纳米材料。我们通过计算设计了假对称异构体成分,并利用它们创建了具有二十面体对称性的离散笼状蛋白质组装体,其中包含 240、540 和 960 个亚基。这些纳米笼的直径分别为 49、71 和 96 纳米,是迄今为止通过计算设计生成的最大的有界蛋白质组装体。更广泛地说,通过超越严格的对称性,我们的工作大大拓宽了可通过设计获得的自组装蛋白质结构的种类。本文章由计算机程序翻译,如有差异,请以英文原文为准。

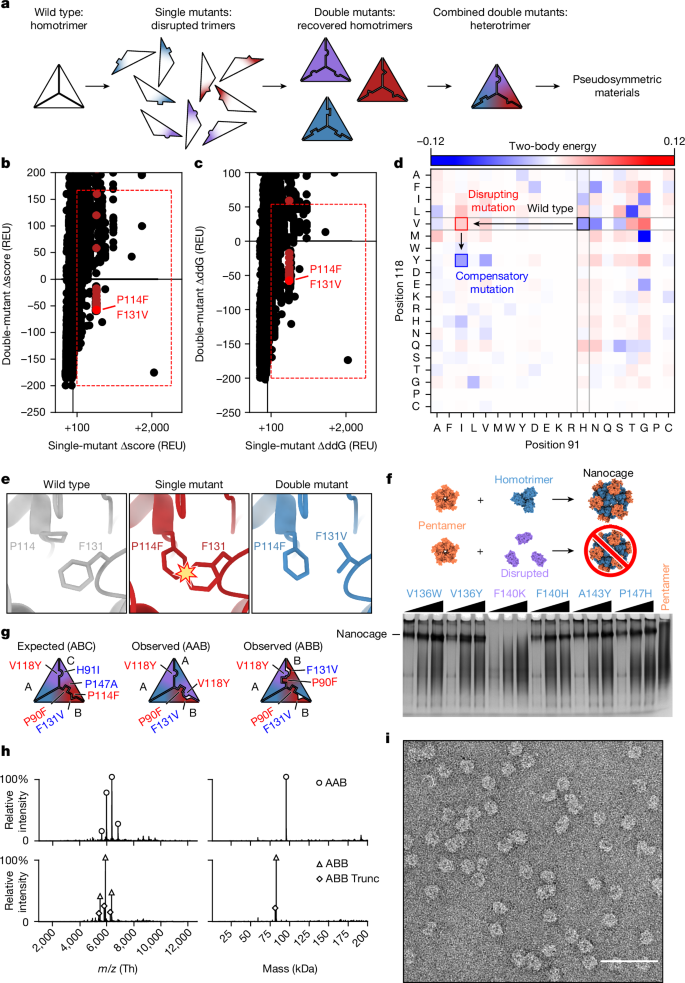
Hierarchical design of pseudosymmetric protein nanocages
Discrete protein assemblies ranging from hundreds of kilodaltons to hundreds of megadaltons in size are a ubiquitous feature of biological systems and perform highly specialized functions1,2. Despite remarkable recent progress in accurately designing new self-assembling proteins, the size and complexity of these assemblies has been limited by a reliance on strict symmetry3. Here, inspired by the pseudosymmetry observed in bacterial microcompartments and viral capsids, we developed a hierarchical computational method for designing large pseudosymmetric self-assembling protein nanomaterials. We computationally designed pseudosymmetric heterooligomeric components and used them to create discrete, cage-like protein assemblies with icosahedral symmetry containing 240, 540 and 960 subunits. At 49, 71 and 96 nm diameter, these nanocages are the largest bounded computationally designed protein assemblies generated to date. More broadly, by moving beyond strict symmetry, our work substantially broadens the variety of self-assembling protein architectures that are accessible through design. A new computational method for design of pseudosymmetric self-assembling protein nanomaterials has resulted in purification of cage-like protein assemblies containing 960 subunits with a diameter of 96 nm.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


