RNase P RNA 在溶液中的构象空间
IF 48.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
RNA 的构象多样性具有基本的生物学作用1,2,3,4,5,但使用传统的生物物理技术无法直接观察其在溶液中的完整构象空间。通过使用溶液原子力显微镜、深度神经网络和统计分析,我们发现核糖核酸酶 P(RNase P)RNA 采用异质构象,包括构象不变的核心和高度灵活的外围结构元素,这些元素可以在广阔的构象空间中采样,在多个方向上的振幅高达 20-60 Å,净能量消耗非常低。增加 Mg2+ 可能会缩小构象空间,从而推动压实并增强酶活性。此外,对空间灵活性和序列保持之间的相关性和反相关性的分析表明,关键区域的结构和动力学的功能作用都包含在主序列中。这些发现揭示了 RNase P 的 RNA 成分中酶精确性和底物杂合性的结构动力学基础。本文章由计算机程序翻译,如有差异,请以英文原文为准。
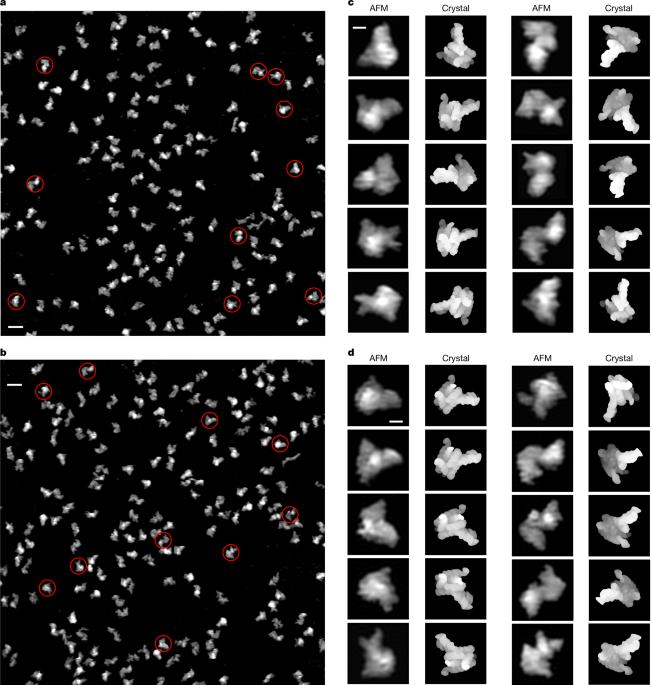
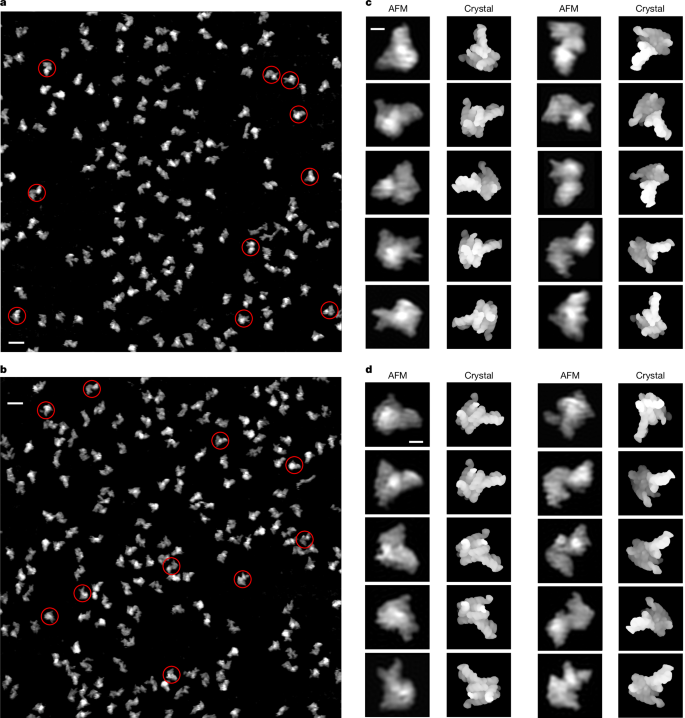
The conformational space of RNase P RNA in solution
RNA conformational diversity has fundamental biological roles1–5, but direct visualization of its full conformational space in solution has not been possible using traditional biophysical techniques. Using solution atomic force microscopy, a deep neural network and statistical analyses, we show that the ribonuclease P (RNase P) RNA adopts heterogeneous conformations consisting of a conformationally invariant core and highly flexible peripheral structural elements that sample a broad conformational space, with amplitudes as large as 20–60 Å in a multitude of directions, with very low net energy cost. Increasing Mg2+ drives compaction and enhances enzymatic activity, probably by narrowing the conformational space. Moreover, analyses of the correlations and anticorrelations between spatial flexibility and sequence conservation suggest that the functional roles of both the structure and dynamics of key regions are embedded in the primary sequence. These findings reveal the structure–dynamics basis for the embodiment of both enzymatic precision and substrate promiscuity in the RNA component of the RNase P. Mapping the conformational space of the RNase P RNA demonstrates a new general approach to studying RNA structure and dynamics. Using a deep neural network and statistical analyses of atomic force microscopy images of individual RNA molecules enables the mapping of RNA conformational space in solution.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


