发育中的小鼠大脑的单细胞细胞计数图谱
IF 20
1区 医学
Q1 NEUROSCIENCES
引用次数: 0
摘要
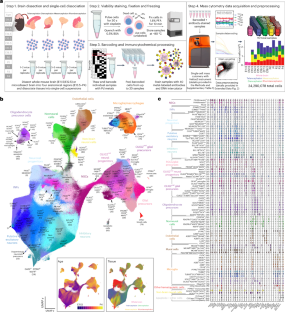
哺乳动物大脑的发育需要不同细胞系之间发生精确的分子变化。虽然单细胞 RNA 测序(scRNA-seq)已表征了发育中大脑的单细胞 RNA 丰度,但单细胞蛋白质丰度尚未表征。为了填补这一空白,我们对 C57/BL6 小鼠胚胎 11.5-E12.5 天的全脑以及出生后 E13.5-Postnatal Day (P)4 的端脑、间脑和菱脑进行了质谱细胞计数。使用 40 种抗体面板分析每个样本中 2 到 4 个生物重复的 24,290,787 个细胞,我们确定了来自不同系的 85 个分子上不同的细胞群。我们的分析证实了神经发生和胶质细胞发生的典型分子途径,并预测了皮质少突发生的两种不同轨迹。经免疫组化和 RNA 原位杂交(ISH)验证,质粒细胞术和 scRNA-seq 得出的蛋白质表达与 RNA 表达的差异,证明了蛋白质水平测量在确定细胞功能状态方面的价值。我们的研究结果表明,质谱仪是一种可扩展的脑组织单细胞分析平台。本文章由计算机程序翻译,如有差异,请以英文原文为准。

A single-cell mass cytometry-based atlas of the developing mouse brain
Development of the mammalian brain requires precise molecular changes across diverse cell lineages. While single-cell RNA abundances in the developing brain have been characterized by single-cell RNA sequencing (scRNA-seq), single-cell protein abundances have not been characterized. To address this gap, we performed mass cytometry on the whole brain at embryonic day (E)11.5–E12.5 and the telencephalon, the diencephalon, the mesencephalon and the rhombencephalon at E13.5–postnatal day (P)4 from C57/BL6 mice. Using a 40-antibody panel to analyze 24,290,787 cells from two to four biological replicates per sample, we identify 85 molecularly distinct cell clusters from distinct lineages. Our analyses confirm canonical molecular pathways of neurogenesis and gliogenesis, and predict two distinct trajectories for cortical oligodendrogenesis. Differences in protein versus RNA expression from mass cytometry and scRNA-seq, validated by immunohistochemistry and RNAscope in situ hybridization (ISH), demonstrate the value of protein-level measurements for identifying functional cell states. Our findings show the utility of mass cytometry as a scalable platform for single-cell profiling of brain tissues. In recent years, valuable mRNA-based developmental atlases of the mouse brain have been made available. Here, the authors use single-cell mass cytometry to build a protein-based single-cell profiling of the developing embryonic and postnatal mouse brain.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature neuroscience
医学-神经科学
CiteScore
38.60
自引率
1.20%
发文量
212
审稿时长
1 months
期刊介绍:
Nature Neuroscience, a multidisciplinary journal, publishes papers of the utmost quality and significance across all realms of neuroscience. The editors welcome contributions spanning molecular, cellular, systems, and cognitive neuroscience, along with psychophysics, computational modeling, and nervous system disorders. While no area is off-limits, studies offering fundamental insights into nervous system function receive priority.
The journal offers high visibility to both readers and authors, fostering interdisciplinary communication and accessibility to a broad audience. It maintains high standards of copy editing and production, rigorous peer review, rapid publication, and operates independently from academic societies and other vested interests.
In addition to primary research, Nature Neuroscience features news and views, reviews, editorials, commentaries, perspectives, book reviews, and correspondence, aiming to serve as the voice of the global neuroscience community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


