质子增强荧光揭示的单分子多价相互作用
IF 15.8
1区 材料科学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
摘要
多价性作为一种相互作用原理在自然界中被广泛应用。它通过增强的亲和力使多种弱相互作用产生特异性强结合,是免疫识别和细胞信号传导的核心过程,也是当前药物设计中的一个概念。在这里,我们利用纳米粒子质子增强荧光的高信号,在单分子水平上实时提取多价相互作用的结合动力学和动态。我们以 DNA 霍利迪接合点(Holliday Junction)为具有可编程价态的模型结构,研究了单价、双价和三价的结合相互作用,并引入了与结构化大分子相关的阶跃结合动力学模型,包括一个可通过实验提取的结合限制项 ω,以量化构象、立体效应和刚性的影响。我们利用这种方法探讨了 DNA 配体的长度和灵活性如何影响结合限制和结合强度,其中整体结合强度随间隔长度的增加而降低。对于三价系统,增加间隔长度会额外激活三价状态下的结合,从而为多价药物或靶向分子的设计提供启示。通过系统地改变受体密度,我们在单分子水平上探索了多价 HJ 的结合超选择性。我们发现三价结合强度的多项式行为清楚地表明了受体密度依赖性选择性结合。有趣的是,我们可以利用等离子体快速衰减的近场,这种近场会引起信号与染料位置的强烈依赖性,从而观察单个多价结合事件中的结合动态。本文章由计算机程序翻译,如有差异,请以英文原文为准。
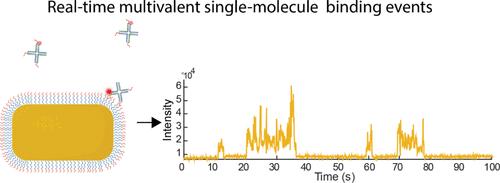
Single-Molecule Multivalent Interactions Revealed by Plasmon-Enhanced Fluorescence
Multivalency as an interaction principle is widely utilized in nature. It enables specific and strong binding by multiple weak interactions through enhanced avidity and is a core process in immune recognition and cellular signaling, which is also a current concept in drug design. Here, we use the high signals from plasmon-enhanced fluorescence of nanoparticles to extract binding kinetics and dynamics of multivalent interactions on the single-molecule level and in real time. We study mono-, bi-, and trivalent binding interactions using a DNA Holliday Junction as a model construct with programmable valency and introduce a step-binding model for binding kinetics relevant for structured macromolecules by including an experimentally extractable binding restriction term ω to quantify the effects from conformation, steric effects, and rigidity. We used this approach to explore how length and flexibility of the DNA ligands affect binding restriction and binding strength, where the overall binding strength decreased with spacer length. For trivalent systems, increasing spacer length additionally activated binding in the trivalent state, giving insight into the design of multivalent drug or targeting moieties. By systematically changing the receptor density, we explored the binding super selectivity of the multivalent HJ at the single-molecule level. We find a polynomial behavior of the trivalent binding strength that clearly shows receptor-density-dependent selective binding. Interestingly, we could exploit the rapidly decaying near fields of the plasmon that induce a strong dependence of the signal on the position of the dye to observe binding dynamics during single multivalent binding events.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

ACS Nano
工程技术-材料科学:综合
CiteScore
26.00
自引率
4.10%
发文量
1627
审稿时长
1.7 months
期刊介绍:
ACS Nano, published monthly, serves as an international forum for comprehensive articles on nanoscience and nanotechnology research at the intersections of chemistry, biology, materials science, physics, and engineering. The journal fosters communication among scientists in these communities, facilitating collaboration, new research opportunities, and advancements through discoveries. ACS Nano covers synthesis, assembly, characterization, theory, and simulation of nanostructures, nanobiotechnology, nanofabrication, methods and tools for nanoscience and nanotechnology, and self- and directed-assembly. Alongside original research articles, it offers thorough reviews, perspectives on cutting-edge research, and discussions envisioning the future of nanoscience and nanotechnology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


