对 18 种人类癌症的基因表达进行深度剖析
IF 26.8
1区 医学
Q1 ENGINEERING, BIOMEDICAL
引用次数: 0
摘要
通过无监督深度学习,可以挖掘大型癌症基因表达数据集中的临床和生物学信息。然而,与生物学可解释性和方法论稳健性相关的困难使得这种方法不切实际。在这里,我们描述了一种无监督深度学习框架,用于为来自 18 种人类癌症的 50,211 个转录组的基因表达数据生成低维潜在空间。我们将这一框架命名为 DeepProfile,它在生物可解释性方面优于降维方法,并使我们得以揭示,在定义不同癌症类型的潜空间时,普遍重要的基因控制着免疫细胞的激活,而癌症类型特异性基因和通路则定义了分子疾病亚型。通过将 DeepProfile 中的潜变量与肿瘤的次要特征联系起来,我们发现突变负荷与细胞周期相关基因的表达密切相关,而 DNA 错配修复和 MHC II 类抗原呈递生物通路的活性与患者的存活率始终相关。我们还发现,肿瘤相关巨噬细胞是与生存相关的 MHC II 类转录本的来源。无监督学习有助于从基因表达数据中发现生物学见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。

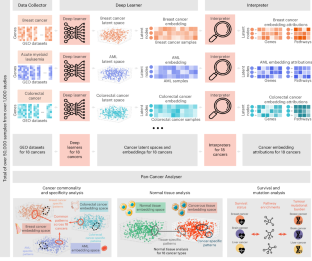
Deep profiling of gene expression across 18 human cancers
Clinical and biological information in large datasets of gene expression across cancers could be tapped with unsupervised deep learning. However, difficulties associated with biological interpretability and methodological robustness have made this impractical. Here we describe an unsupervised deep-learning framework for the generation of low-dimensional latent spaces for gene-expression data from 50,211 transcriptomes across 18 human cancers. The framework, which we named DeepProfile, outperformed dimensionality-reduction methods with respect to biological interpretability and allowed us to unveil that genes that are universally important in defining latent spaces across cancer types control immune cell activation, whereas cancer-type-specific genes and pathways define molecular disease subtypes. By linking latent variables in DeepProfile to secondary characteristics of tumours, we discovered that mutation burden is closely associated with the expression of cell-cycle-related genes, and that the activity of biological pathways for DNA-mismatch repair and MHC class II antigen presentation are consistently associated with patient survival. We also found that tumour-associated macrophages are a source of survival-correlated MHC class II transcripts. Unsupervised learning can facilitate the discovery of biological insight from gene-expression data. Using unsupervised deep learning to generate low-dimensional latent spaces for gene-expression data can unveil biological insight across cancers.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Biomedical Engineering
Medicine-Medicine (miscellaneous)
CiteScore
45.30
自引率
1.10%
发文量
138
期刊介绍:
Nature Biomedical Engineering is an online-only monthly journal that was launched in January 2017. It aims to publish original research, reviews, and commentary focusing on applied biomedicine and health technology. The journal targets a diverse audience, including life scientists who are involved in developing experimental or computational systems and methods to enhance our understanding of human physiology. It also covers biomedical researchers and engineers who are engaged in designing or optimizing therapies, assays, devices, or procedures for diagnosing or treating diseases. Additionally, clinicians, who make use of research outputs to evaluate patient health or administer therapy in various clinical settings and healthcare contexts, are also part of the target audience.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


