使用PASTE在哺乳动物细胞中精确插入千碱基规模的基因组。
IF 16
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
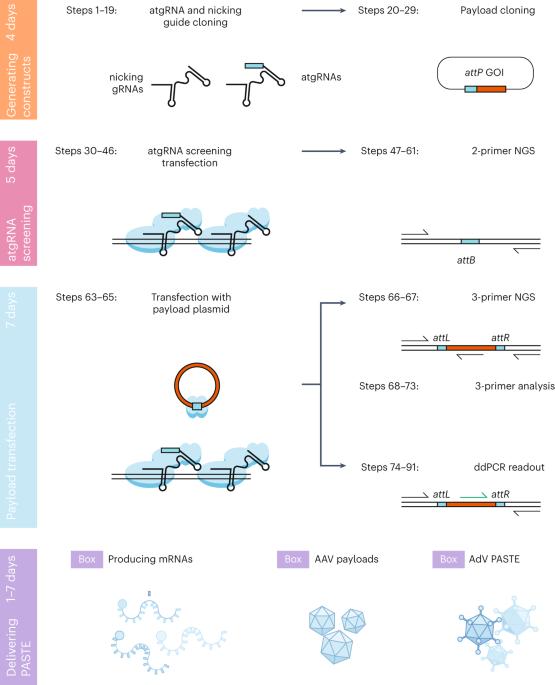
可编程基因整合技术是一种新兴模式,在基础研究和治疗开发方面都有令人兴奋的应用。通过位点特异性靶向元件进行可编程添加(PASTE)是一种可编程基因整合方法,用于将大型 DNA 序列精确、高效地可编程整合到基因组中。与以往将长序列插入哺乳动物基因组的方法相比,PASTE 提高了编辑效率、纯度和可编程性。PASTE 将位点特异性整合酶的特异性和货物大小能力与质粒编辑的可编程性相结合,可以精确插入至少 36 kb 的货物,效率高达 60%。在此,我们概述了设计、执行和分析 PASTE 实验的最佳实践,包括在人类 NOLC1 和 ACTB 基因组位点整合 EGFP 的方案,以及通过新一代测序和液滴数字 PCR 进行读出的方案。我们提供了设计和优化定制 PASTE 实验的指南,以便在其他基因组位点整合所需的有效载荷,还提供了框内蛋白标记和多重插入的应用实例。为方便实验设置,我们提供了必要的序列和质粒,以便通过质粒转染或体外转录 RNA 将 PASTE 成分输送到细胞中。本方案中的大多数实验可在短短两周内完成,从而实现精确、多用途的可编程基因插入。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Precise kilobase-scale genomic insertions in mammalian cells using PASTE
Programmable gene integration technologies are an emerging modality with exciting applications in both basic research and therapeutic development. Programmable addition via site-specific targeting elements (PASTE) is a programmable gene integration approach for precise and efficient programmable integration of large DNA sequences into the genome. PASTE offers improved editing efficiency, purity and programmability compared with previous methods for long insertions into the mammalian genome. By combining the specificity and cargo size capabilities of site-specific integrases with the programmability of prime editing, PASTE can precisely insert cargoes of at least 36 kb with efficiencies of up to 60%. Here we outline best practices for design, execution and analysis of PASTE experiments, with protocols for integration of EGFP at the human NOLC1 and ACTB genomic loci and for readout by next generation sequencing and droplet digital PCR. We provide guidelines for designing and optimizing a custom PASTE experiment for integration of desired payloads at alternative genomic loci, as well as example applications for in-frame protein tagging and multiplexed insertions. To facilitate experimental setup, we include the necessary sequences and plasmids for the delivery of PASTE components to cells via plasmid transfection or in vitro transcribed RNA. Most experiments in this protocol can be performed in as little as 2 weeks, allowing for precise and versatile programmable gene insertion. Programmable addition via site-specific targeting elements (PASTE) combines the specificity, efficiency and cargo size benefits of site-specific integrases with the programmability of prime editing for precise and efficient integration of large DNA sequences into mammalian genomes.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


