利用有利单倍型优化株高提高水稻产量和耐盐性
IF 13
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
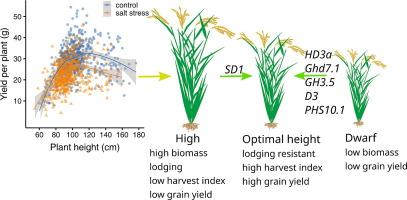
水稻(Oryza sativa L.)是全球数十亿人的主食,目前正面临盐胁迫的挑战。由于对水稻耐盐生理和遗传基础的认识有限,很少有基因被确定为水稻育种的有价值基因,这是水稻高产耐盐品种开发的主要瓶颈。目的筛选具有育种潜力的水稻耐盐基因/数量性状位点。方法采用166个盐渍化地区的中国水稻品种和412个国际水稻品种进行耐盐性田间试验。采用全基因组关联研究(GWAS)鉴定了与高产和耐盐相关的关键位点。此外,还评估了引入有益单倍型对籽粒产量和耐盐性的影响。结果水稻在正常和盐胁迫条件下保持高产的最佳株高为100 ~ 120 cm。GWAS发现了6个与不同环境下水稻生长和籽粒产量相关的新qtl /基因,不同于之前发现的盐胁迫相关基因。值得注意的是,编码丝氨酸/苏氨酸蛋白激酶的基因PHS10.1可能调节碳代谢、淀粉和蔗糖代谢,影响植物生长和粮食产量。传统育种选择了SD1、Ghd7.1、GH3.5和PHS10.1等调节株高和籽粒产量的基因单倍型。此外,通过导入这些基因的有益等位基因来优化株高,在正常和盐水条件下均可提高受体的产量。结论利用有益单倍型来优化株高可以有效地平衡水稻生长与胁迫之间的平衡。这代表了一种很有前途的育种策略,可以开发出既高产又耐盐的作物品种。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Improving grain yield and salt tolerance by optimizing plant height with beneficial haplotypes in rice (Oryza sativa)
Introduction
Rice (Oryza sativa L.), a staple food for billions worldwide, is challenged by salt stress. Owing to the limited understanding of the physiological and genetic basis of rice salt tolerance, few genes have been identified as valuable in rice breeding, causing a major bottleneck in the development of high-yield, salt-tolerant rice varieties.
Objective
This study aims to identify salt tolerance genes/quantitative trait loci (QTLs) with breeding potential in rice.
Methods
Field trials were conducted with 166 Chinese rice cultivars from saline-affected regions and 412 global rice accessions to assess salt tolerance. Genome-wide association study (GWAS) was performed to identify key loci related to high yield and salt tolerance. Additionally, the impact of introducing beneficial haplotypes on grain yield and salt tolerance was assessed.
Results
The optimal rice plant height of 100–120 cm was crucial for sustaining high yield under both normal and salt stress conditions. GWAS revealed 6 novel QTLs/genes associated with rice plant growth and grain yield across various environments, distinct from previously recognized salt stress-related genes. Notably, the gene PHS10.1, encoding a serine/threonine protein kinase, may regulate carbon metabolism, starch and sucrose metabolism, influencing plant growth and grain yield. Certain haplotypes of the genes regulating plant height and grain yield, including SD1, Ghd7.1, GH3.5, and PHS10.1, were selected in traditional breeding. Moreover, optimizing plant height through the introgression of beneficial alleles of these genes increased grain yield in recipient lines under both normal and saline conditions.
Conclusion
We propose that utilizing beneficial haplotypes to optimize plant height can effectively balance the growth–stress trade-offs in rice plants. This represents a promising breeding strategy for the development of crop varieties that are both high-yielding and salt-tolerant.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Advanced Research
Multidisciplinary-Multidisciplinary
CiteScore
21.60
自引率
0.90%
发文量
280
审稿时长
12 weeks
期刊介绍:
Journal of Advanced Research (J. Adv. Res.) is an applied/natural sciences, peer-reviewed journal that focuses on interdisciplinary research. The journal aims to contribute to applied research and knowledge worldwide through the publication of original and high-quality research articles in the fields of Medicine, Pharmaceutical Sciences, Dentistry, Physical Therapy, Veterinary Medicine, and Basic and Biological Sciences.
The following abstracting and indexing services cover the Journal of Advanced Research: PubMed/Medline, Essential Science Indicators, Web of Science, Scopus, PubMed Central, PubMed, Science Citation Index Expanded, Directory of Open Access Journals (DOAJ), and INSPEC.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


