ashwagandha (Withania somnifera (L.))多样性和种群结构的遗传分析利用EST-SSR、ISSR和SSR标记:提高农业和工业价值的意义
IF 5.6
1区 农林科学
Q1 AGRICULTURAL ENGINEERING
引用次数: 0
摘要
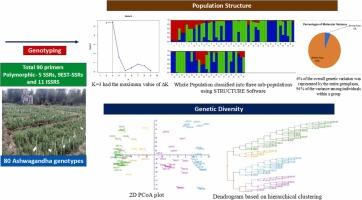
尽管越来越多的人喜欢草药而不是合成产品,但由于对其遗传多样性的研究有限,市场对重要的阿育吠陀草药阿什瓦根达的需求尚未得到满足。本研究利用90个引物,包括简单序列重复序列、表达序列标签- ssrs和简单序列重复序列,对80个不同的Ashwagandha基因型的遗传多样性和群体结构进行了评估,从而弥补了这一空白。该研究证实了19个ssr的交叉可转移性,强调了它们在多样性分析中的未来应用。共得到133个等位基因,平均每个引物有5.32个等位基因。UBC 826(0.89, 13.15)和EMS-12(0.87, 10.88)分别具有最高的多态性信息含量和分辨能力。这些标记具有标记指数最高的特点,有助于鉴定独特的遗传谱,从而有效地选择遗传多样性和优良的育种品系。EMS-12与类异戊二烯生物合成有关,在糖醇内酯的生产中起着至关重要的作用。因此,该引物可以整合到ashwagandha遗传图谱中,以帮助鉴定与次生代谢物产生相关的数量性状位点。值得注意的是,目前还没有研究揭示这种强大草药的遗传种群动态,这促使我们进行这项研究,以进一步探索其含义。本研究共鉴定出3个亚种群,其中种群2的香农信息指数最高,表明其遗传丰富度较高。AMOVA的结果显示了大量的种群内多样性,占群体内总遗传变异的94% %。通过观察2号群体的基因型,进一步证实了这种多样性。此后,树状图和主成分分析通过一致的聚类模式证实了STRUCTURE结果。本研究揭示了Ashwagandha群体的遗传丰富度,为选择和培育具有最佳多样性和理想性状的基因型提供了路线图。这些发现在满足工业和制药需求方面具有巨大的潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Genetic insights into diversity and population structure of ashwagandha (Withania somnifera (L.) Dunal) using EST-SSR, ISSR and SSR markers: Implications for enhancing agricultural and industrial value
Despite rising preference for herbal remedies over synthetic products, market demand for the crucial Ayurvedic herb Ashwagandha is unmet due to limited research on its genetic diversity. This study addresses this gap by utilizing 90 primers including Simple Sequence Repeats, Expressed Sequence Tags-SSRs and Inter Simple Sequence Repeats to assess the genetic diversity and population structure of 80 diverse Ashwagandha genotypes. The study confirmed the cross-transferability of 19 SSRs underscoring their future utility in diversity analysis. A total of 133 alleles were generated with an average of 5.32 alleles per primer. UBC 826 (0.89, 13.15) and EMS-12 (0.87, 10.88) were notable for their highest polymorphic information content and resolving power, respectively. These markers, characterized by the highest marker index, facilitated the identification of unique genetic profiles essential for selecting genetically diverse and superior breeding lines effectively. EMS-12, linked to isoprenoid biosynthesis, plays a crucial role in withanolide production. Thus, this primer can be incorporated into genetic maps of ashwagandha to help identify quantitative trait loci associated with secondary metabolite production. Remarkably, no studies have yet unraveled the genetic population dynamics of this powerful herb, prompting us to undertake this study to explore its implications further. Three subpopulations were identified in the current research, with Population 2 exhibiting the highest Shannon's Information Index, underscoring its genetic richness. The results of AMOVA revealed substantial intra-population diversity, with 94 % of the total genetic variance within groups. This diversity was further confirmed by observing genotypes from population 2 clustered into two distinct groups. Henceforth, dendrogram and principal component analyses confirmed the STRUCTURE results through consistent clustering patterns. This study uncovers genetic richness in Ashwagandha populations, offering a roadmap for selecting and breeding genotypes with optimal diversity and desirable traits to enhance cultivation. The findings hold significant potential for meeting industrial and pharmaceutical needs.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Industrial Crops and Products
农林科学-农业工程
CiteScore
9.50
自引率
8.50%
发文量
1518
审稿时长
43 days
期刊介绍:
Industrial Crops and Products is an International Journal publishing academic and industrial research on industrial (defined as non-food/non-feed) crops and products. Papers concern both crop-oriented and bio-based materials from crops-oriented research, and should be of interest to an international audience, hypothesis driven, and where comparisons are made statistics performed.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


