CRISPR/Cas13a 与反转录和 RPA 结合用于监测水环境中的 NoV GII.4
IF 10.3
1区 环境科学与生态学
Q1 ENVIRONMENTAL SCIENCES
引用次数: 0
摘要
本文章由计算机程序翻译,如有差异,请以英文原文为准。
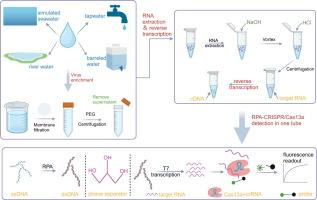
CRISPR/Cas13a combined with reverse transcription and RPA for NoV GII.4 monitoring in water environments
Water bodies contaminated with the norovirus (NoV) are important vectors for its transmission. Therefore, enhanced monitoring of NoV in aqueous environments plays an active role in preventing diseases. Here, we reverse transcribed viral RNA into cDNA, and then used the constructed RPA-CRISPR/Cas13a-based platform for sensitive and quantitative monitoring of NoV GII.4 in aqueous environments. The use of glycerol as a phase separator and the direct release of nucleic acids from the virus by NaOH significantly enhanced the stability of the assay and reduced its economic cost. This assay is sensitive, specific, and stable. Based on the qualitative detection method, we established a relatively accurate quantitative detection method using the plasmid as a standard. Four water samples, totaling 64 samples, were analyzed using this method and compared with the qPCR method. The results of the two methods showed 100 % concordance with no significant difference in viral load. The entire process of our established method—from viral nucleic acid extraction to the output of the results—was completed in 30 min, much less than the time required for qPCR method. This suggests that the assay can be used as an alternative to qPCR for monitoring the change of NoV GII.4 concentration in water bodies, and shows high potential for application in the immediate detection of viruses in aqueous environments and resource-limited areas.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Environment International
环境科学-环境科学
CiteScore
21.90
自引率
3.40%
发文量
734
审稿时长
2.8 months
期刊介绍:
Environmental Health publishes manuscripts focusing on critical aspects of environmental and occupational medicine, including studies in toxicology and epidemiology, to illuminate the human health implications of exposure to environmental hazards. The journal adopts an open-access model and practices open peer review.
It caters to scientists and practitioners across all environmental science domains, directly or indirectly impacting human health and well-being. With a commitment to enhancing the prevention of environmentally-related health risks, Environmental Health serves as a public health journal for the community and scientists engaged in matters of public health significance concerning the environment.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


