排列CRISPR文库,用于人类蛋白质编码基因的全基因组激活、缺失和沉默
IF 26.8
1区 医学
Q1 ENGINEERING, BIOMEDICAL
引用次数: 0
摘要
排列的CRISPR文库将基因扰动筛选的范围扩展到非选择性细胞表型。然而,文库的生成需要组装数千个表达单导rna (sgRNAs)的载体。在这里,利用大规模平行质粒克隆方法,我们发现阵列文库可以用于人类蛋白质编码基因的全基因组消蚀(19,936个质粒)和它们的激活和表观遗传沉默(22,442个质粒),每个质粒编码一个由四个非重叠的sgrna组成的阵列,旨在容忍大多数人类DNA多态性。四倍sgrna文库在缺失(75-99%)和沉默(76-92%)实验中具有较高的扰动效率,在激活实验中具有显著的折叠变化。此外,1634个人类转录因子的排列激活筛选发现了11个细胞朊病毒蛋白PrPC的新调节因子,使用烧蚀库的汇总版本筛选导致鉴定了5个新的自噬修饰因子,否则无法检测到,并且“后汇总”单独产生的慢病毒消除了模板切换伪影并增强了汇总筛选的表观遗传沉默性能。四倍sgrna阵列文库是靶向全基因组扰动的强大而通用的资源。本文章由计算机程序翻译,如有差异,请以英文原文为准。
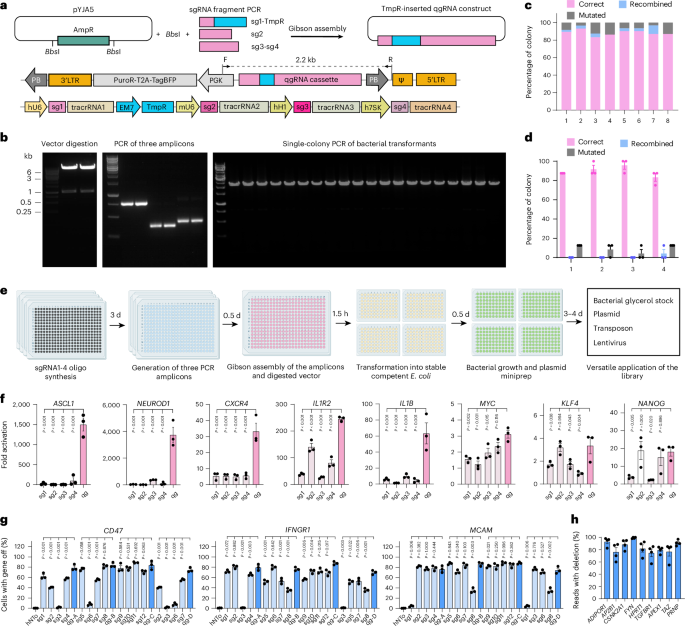

Arrayed CRISPR libraries for the genome-wide activation, deletion and silencing of human protein-coding genes
Arrayed CRISPR libraries extend the scope of gene-perturbation screens to non-selectable cell phenotypes. However, library generation requires assembling thousands of vectors expressing single-guide RNAs (sgRNAs). Here, by leveraging massively parallel plasmid-cloning methodology, we show that arrayed libraries can be constructed for the genome-wide ablation (19,936 plasmids) of human protein-coding genes and for their activation and epigenetic silencing (22,442 plasmids), with each plasmid encoding an array of four non-overlapping sgRNAs designed to tolerate most human DNA polymorphisms. The quadruple-sgRNA libraries yielded high perturbation efficacies in deletion (75–99%) and silencing (76–92%) experiments and substantial fold changes in activation experiments. Moreover, an arrayed activation screen of 1,634 human transcription factors uncovered 11 novel regulators of the cellular prion protein PrPC, screening with a pooled version of the ablation library led to the identification of 5 novel modifiers of autophagy that otherwise went undetected, and ‘post-pooling’ individually produced lentiviruses eliminated template-switching artefacts and enhanced the performance of pooled screens for epigenetic silencing. Quadruple-sgRNA arrayed libraries are a powerful and versatile resource for targeted genome-wide perturbations. Arrayed CRISPR libraries with each plasmid encoding an array of four non-overlapping single-guide RNAs allow for the efficient genome-wide ablation, activation and epigenetic silencing of human protein-coding genes.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Biomedical Engineering
Medicine-Medicine (miscellaneous)
CiteScore
45.30
自引率
1.10%
发文量
138
期刊介绍:
Nature Biomedical Engineering is an online-only monthly journal that was launched in January 2017. It aims to publish original research, reviews, and commentary focusing on applied biomedicine and health technology. The journal targets a diverse audience, including life scientists who are involved in developing experimental or computational systems and methods to enhance our understanding of human physiology. It also covers biomedical researchers and engineers who are engaged in designing or optimizing therapies, assays, devices, or procedures for diagnosing or treating diseases. Additionally, clinicians, who make use of research outputs to evaluate patient health or administer therapy in various clinical settings and healthcare contexts, are also part of the target audience.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


