倒置对沙丘向日葵平行基因组分化的贡献不成比例
IF 13.9
1区 生物学
Q1 ECOLOGY
引用次数: 0
摘要
平行遗传进化的可能性是遗传结构所施加的选择强度和约束的函数。逆转录捕获局部适应的等位基因并抑制它们之间的重组,这限制了适应反应的范围。此外,反转内等位基因的组合表型效应可能大于单个等位基因的组合表型效应;这将进一步增加反转对并行进化的贡献。我们验证了倒置在独立沙丘生态型中对平行遗传进化的贡献不成比例的假设。我们分析了栖息地数据,并确定了平行栖息地转移的变量。对这些变量的基因型-环境关联分析表明,对共同选择压力的反转有平行的反应。我们还证实了沙丘的种子大小更大,并通过多个杂交进行了数量性状位点定位。不同位置间数量性状基因座共享发生倒转的概率大于预期。我们使用全基因组测序数据来确定沙丘生态型中的选择性扫描,并发现大多数共享扫描区域都在反转中发现。共同区域的系统发育分析表明,在反转中,在两个地点的沙丘生境中通常发现相同的等位基因。这些结果证实了逆温驱动沙丘生态类型平行分化的预测。本文章由计算机程序翻译,如有差异,请以英文原文为准。

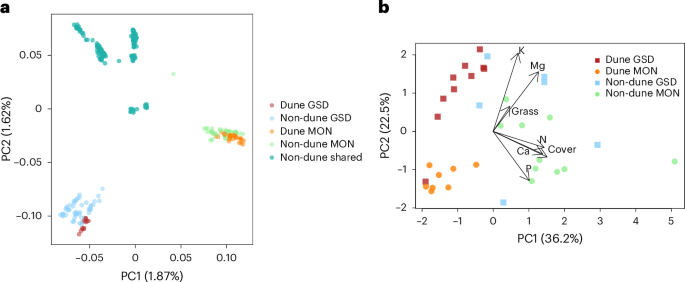
Inversions contribute disproportionately to parallel genomic divergence in dune sunflowers
The probability of parallel genetic evolution is a function of the strength of selection and constraints imposed by genetic architecture. Inversions capture locally adapted alleles and suppress recombination between them, which limits the range of adaptive responses. In addition, the combined phenotypic effect of alleles within inversions is likely to be greater than that of individual alleles; this should further increase the contributions of inversions to parallel evolution. We tested the hypothesis that inversions contribute disproportionately to parallel genetic evolution in independent dune ecotypes of Helianthus petiolaris. We analysed habitat data and identified variables underlying parallel habitat shifts. Genotype–environment association analyses of these variables indicated parallel responses of inversions to shared selective pressures. We also confirmed larger seed size across the dunes and performed quantitative trait locus mapping with multiple crosses. Quantitative trait loci shared between locations fell into inversions more than expected by chance. We used whole-genome sequencing data to identify selective sweeps in the dune ecotypes and found that the majority of shared swept regions were found within inversions. Phylogenetic analyses of shared regions indicated that within inversions, the same allele typically was found in the dune habitat at both sites. These results confirm predictions that inversions drive parallel divergence in the dune ecotypes. Analysis of habitat data, quantitative trait locus mapping of seed size and selective sweeps show parallel selection acting on inversions in two independent dune ecotypes of the prairie sunflower, Helianthus petiolaris.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature ecology & evolution
Agricultural and Biological Sciences-Ecology, Evolution, Behavior and Systematics
CiteScore
22.20
自引率
2.40%
发文量
282
期刊介绍:
Nature Ecology & Evolution is interested in the full spectrum of ecological and evolutionary biology, encompassing approaches at the molecular, organismal, population, community and ecosystem levels, as well as relevant parts of the social sciences. Nature Ecology & Evolution provides a place where all researchers and policymakers interested in all aspects of life's diversity can come together to learn about the most accomplished and significant advances in the field and to discuss topical issues. An online-only monthly journal, our broad scope ensures that the research published reaches the widest possible audience of scientists.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


