使用基因组优先方法研究人类TM6SF2蛋白改变变异的深度代谢表型
IF 9.5
1区 医学
Q1 GASTROENTEROLOGY & HEPATOLOGY
引用次数: 0
摘要
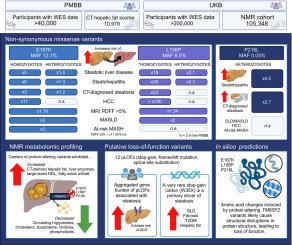
背景,AimAn无偏倚的基因组优先方法可以扩大对疾病不可知论生物库中特定基因的分子理解,从而进行更深入的表型分析。由于已知TM6SF2与脂肪变性肝病(SLD)相关,因此TM6SF2是该方法的良好候选者。方法我们在Penn Medicine Biobank (PMBB, n >40,000)和UK Biobank (UKB, n >200,000)中筛选具有TM6SF2蛋白改变变异的全外显子组序列的参与者,并评估其与肝脏表型和临床结果的关联。结果TM6SF2错义变异(E167K, L156P, P216L)与临床诊断和影像学证实的脂肪变性风险增加相关,独立于PNPLA3 I48M风险等位基因和乙型/丙型肝炎(p <0.001)。E167K纯合子显著增加SLD(比值比5.38,p <0.001)、脂肪性肝炎(比值比5.76,p <0.05)和肝细胞癌(比值比11.22,p <0.0001)的风险,而L156P和P216L杂合携带者患脂肪性肝炎的风险也增加。此外,携带E167K的人患高危MASH的风险增加了3倍(OR 2.75, p <0.001)。在等位基因剂量方面,E167K和L156P的ct肝脂肪评分更高(p <0.05)。这与UKB核磁共振衍生的脂质组学分析相一致(n = 105,348),显示所有携带者都表现出较低的总胆固醇、甘油三酯和总胆碱。计算机预测表明,这些错义变异导致EXPERA结构域的结构破坏,导致蛋白质功能降低。这一假设得到了TM6SF2中罕见的功能丧失变异与SLD风险增加的关联(OR 4.9, p <0.05)的支持,这主要是由一种具有相同方向性的新型罕见的停止增益变异(W35X)驱动的。结论蛋白改变变异的功能遗传学研究为了解TM6SF2功能丧失与SLD之间的关系提供了新的思路,为今后的机制研究提供了基础。影响和启示:基因组优先的方法扩展了对脂肪变性肝病遗传风险因素的见解,由于已知TM6SF2与血浆脂质特征相关,因此成为焦点。我们的研究结果证实了两种错义变异(E167K和L156P)与CT和MRI扫描显示的肝脂肪变性风险增加的关联,以及独立于常见PNPLA3 I48M风险变异的临床诊断的肝细胞癌风险。值得注意的是,我们还发现了一种预测的有害错义变异(P216L)与脂肪变性风险相关,并证明了罕见的假定功能丧失变异的聚集基因负担与肝脏脂肪变性风险相关。综上所述,本研究为未来TM6SF2变异在代谢功能障碍相关脂肪变性肝病中的功能影响机制研究奠定了基础。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Deep metabolic phenotyping of humans with protein-altering variants in TM6SF2 using a genome-first approach
Background & Aim
An unbiased genome-first approach can expand the molecular understanding of specific genes in disease-agnostic biobanks for deeper phenotyping. TM6SF2 represents a good candidate for this approach due to its known association with steatotic liver disease (SLD).
Methods
We screened participants with whole-exome sequences in the Penn Medicine Biobank (PMBB, n >40,000) and the UK Biobank (UKB, n >200,000) for protein-altering variants in TM6SF2 and evaluated their association with liver phenotypes and clinical outcomes.
Results
Missense variants in TM6SF2 (E167K, L156P, P216L) were associated with an increased risk of clinically diagnosed and imaging-proven steatosis, independent of the PNPLA3 I48M risk allele and hepatitis B/C (p <0.001). E167K homozygotes had significantly increased risk of SLD (odds ratio [OR] 5.38, p <0.001), steatohepatitis (OR 5.76, p <0.05) and hepatocellular carcinoma (OR 11.22, p <0.0001), while heterozygous carriers of L156P and P216L were also at an increased risk of steatohepatitis. In addition, carriers of E167K are at a 3-fold increased risk of at-risk MASH (OR 2.75, p <0.001). CT-derived liver fat scores were higher in E167K and L156P in an allele-dose manner (p <0.05). This corresponded with the UKB nuclear magnetic resonance-derived lipidomic analyses (n = 105,348), revealing all carriers to exhibit lower total cholesterol, triglycerides and total choline. In silico predictions suggested that these missense variants cause structural disruptions in the EXPERA domain, leading to reduced protein function. This hypothesis was supported by the association of rare loss-of-function variants in TM6SF2 with an increased risk of SLD (OR 4.9, p <0.05), primarily driven by a novel rare stop-gain variant (W35X) with the same directionality.
Conclusion
The functional genetic study of protein-altering variants provides insights on the association between loss of TM6SF2 function and SLD and provides the basis for future mechanistic studies.
Impact and implications:
The genome-first approach expands insights into genetic risk factors for steatotic liver disease with TM6SF2 being a focal point due to its known association with plasma lipid traits. Our findings validated the association of two missense variants (E167K and L156P) with increased risk of hepatic steatosis on CT and MRI scans, as well as the risk of clinically diagnosed hepatocellular carcinoma independent of the common PNPLA3 I48M risk variant. Notably, we also identified a predicted deleterious missense variant (P216L) linked to steatotic risk and demonstrated that an aggregated gene burden of rare putative loss-of-function variants was associated with the risk of hepatic steatosis. Combined, this study sets the stage for future mechanistic investigations into the functional consequences of TM6SF2 variants in metabolic dysfunction-associated steatotic liver disease.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

JHEP Reports
GASTROENTEROLOGY & HEPATOLOGY-
CiteScore
12.40
自引率
2.40%
发文量
161
审稿时长
36 days
期刊介绍:
JHEP Reports is an open access journal that is affiliated with the European Association for the Study of the Liver (EASL). It serves as a companion journal to the highly respected Journal of Hepatology.
The primary objective of JHEP Reports is to publish original papers and reviews that contribute to the advancement of knowledge in the field of liver diseases. The journal covers a wide range of topics, including basic, translational, and clinical research. It also focuses on global issues in hepatology, with particular emphasis on areas such as clinical trials, novel diagnostics, precision medicine and therapeutics, cancer research, cellular and molecular studies, artificial intelligence, microbiome research, epidemiology, and cutting-edge technologies.
In summary, JHEP Reports is dedicated to promoting scientific discoveries and innovations in liver diseases through the publication of high-quality research papers and reviews covering various aspects of hepatology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


