用SPRINTER表征单细胞克隆中肿瘤增殖的进化动力学
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
增殖是癌症的一个关键标志,但在肿瘤内共存的进化上不同的克隆之间是否存在差异尚不清楚。我们通过进化路径(SPRINTER)算法引入非同质肿瘤中的单细胞增殖率推断,该算法使用单细胞全基因组DNA测序数据,通过生成准确的基线数据来评估S期和g2期细胞的准确鉴定和克隆分配。应用于新生成的14,994个非小细胞肺癌细胞的纵向,原发性转移匹配数据集,SPRINTER显示了广泛的克隆增殖异质性,Ki-67染色,细胞核成像和临床影像学正交支持。我们进一步证明,高增殖克隆具有更高的转移播种潜力,增加循环肿瘤DNA脱落和克隆特异性改变增殖或转移相关基因的复制时间,这些基因与表达变化相关。应用于先前生成的61,914个乳腺癌和卵巢癌细胞的数据集,SPRINTER揭示了不同基因组变异的单细胞率增加,以及高增殖克隆中增殖相关基因扩增的富集。本文章由计算机程序翻译,如有差异,请以英文原文为准。
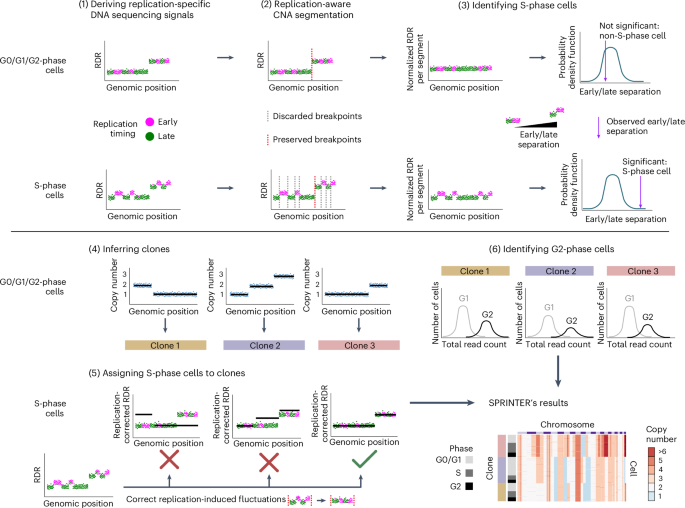

Characterizing the evolutionary dynamics of cancer proliferation in single-cell clones with SPRINTER
Proliferation is a key hallmark of cancer, but whether it differs between evolutionarily distinct clones co-existing within a tumor is unknown. We introduce the Single-cell Proliferation Rate Inference in Non-homogeneous Tumors through Evolutionary Routes (SPRINTER) algorithm that uses single-cell whole-genome DNA sequencing data to enable accurate identification and clone assignment of S- and G2-phase cells, as assessed by generating accurate ground truth data. Applied to a newly generated longitudinal, primary-metastasis-matched dataset of 14,994 non-small cell lung cancer cells, SPRINTER revealed widespread clone proliferation heterogeneity, orthogonally supported by Ki-67 staining, nuclei imaging and clinical imaging. We further demonstrated that high-proliferation clones have increased metastatic seeding potential, increased circulating tumor DNA shedding and clone-specific altered replication timing in proliferation- or metastasis-related genes associated with expression changes. Applied to previously generated datasets of 61,914 breast and ovarian cancer cells, SPRINTER revealed increased single-cell rates of different genomic variants and enrichment of proliferation-related gene amplifications in high-proliferation clones. Single-cell Proliferation Rate Inference in Non-homogeneous Tumors through Evolutionary Routes (SPRINTER) allows users to infer proliferation rates of individual clones within a tumor from single-cell DNA sequencing data. Applying SPRINTER to human tumor datasets highlighted a link between proliferation and metastatic potential.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


