释放作物的遗传潜力:基因组和表观基因组编辑调控区的进展
IF 7.5
2区 生物学
Q1 PLANT SCIENCES
引用次数: 0
摘要
基因组编辑工具可以精确有效地针对植物基因组进行编辑,从而开发出改良作物。除了编辑编码区,研究人员还可以利用编辑技术针对特定的基因调控元件或修改与远端调控区相关的表观遗传标记,从而调控作物中的基因表达。本综述概述了几种著名的基因组编辑技术,包括 CRISPR-Cas9、TALENs 和 ZFNs 以及最新进展。还讨论了基因组和表观基因组编辑的应用,特别是作物植物中调控区的编辑,包括提高非生物胁迫耐受性、产量、抗病性和植物表型。此外,综述还探讨了表观遗传修饰(如 DNA 甲基化和组蛋白修饰)在改变基因表达以改良作物方面的潜力。最后,还讨论了利用各种基因组编辑工具操纵基因调控元件的局限性和未来范围,以充分释放这些工具在植物育种中的潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
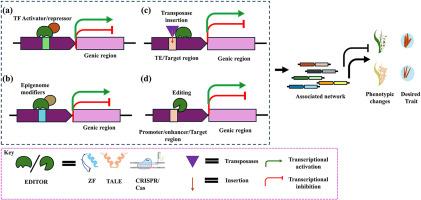
Unlocking crops’ genetic potential: Advances in genome and epigenome editing of regulatory regions
Genome editing tools could precisely and efficiently target plant genomes leading to the development of improved crops. Besides editing the coding regions, researchers can employ editing technologies to target specific gene regulatory elements or modify epigenetic marks associated with distal regulatory regions, thereby regulating gene expression in crops. This review outlines several prominent genome editing technologies, including CRISPR-Cas9, TALENs, and ZFNs and recent advancements. The applications for genome and epigenome editing especially of regulatory regions in crop plants is also discussed, including efforts to enhance abiotic stress tolerance, yield, disease resistance and plant phenotype. Additionally, the review addresses the potential of epigenetic modifications, such as DNA methylation and histone modifications, to alter gene expression for crop improvement. Finally, the limitations and future scope of utilizing various genome editing tools to manipulate regulatory elements for gene regulation to unlock the full potential of these tools in plant breeding has been discussed.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Current opinion in plant biology
生物-植物科学
CiteScore
16.30
自引率
3.20%
发文量
131
审稿时长
6-12 weeks
期刊介绍:
Current Opinion in Plant Biology builds on Elsevier's reputation for excellence in scientific publishing and long-standing commitment to communicating high quality reproducible research. It is part of the Current Opinion and Research (CO+RE) suite of journals. All CO+RE journals leverage the Current Opinion legacy - of editorial excellence, high-impact, and global reach - to ensure they are a widely read resource that is integral to scientists' workflow.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


