基因模块重构确定了斑马鱼的细胞分化过程和特化分泌的调控逻辑
IF 8.7
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
摘要
在分化过程中,细胞会在结构上和功能上变得特化,但人们对其背后的重塑过程却缺乏全面的了解。在这里,我们利用单细胞 RNA 测序(scRNA-seq)发育轨迹,以两种分泌组织--斑马鱼脊索和孵化腺--为模型重建分化。首先,我们整合了表达和功能相似性来识别基因模块,发现了数十个代表已知和新相关分化过程及其动态的模块。其次,我们重点研究了未折叠蛋白反应(UPR)转导模块,以研究一般分泌功能与细胞类型特异性分泌功能是如何调控的。对功能缺失和功能增益胚胎的剖析发现,UPR转录因子creb3l1、creb3l2和xbp1是一般分泌程序的主调控因子,creb3l1/creb3l2还能激活细胞外基质分泌程序,而xbp1则与bhlha15合作激活腺体样分泌程序。我们的研究通过多源整合重构分化(MIMIR)进行了模块识别,并说明了转录因子如何赋予细胞一般和特化功能。本文章由计算机程序翻译,如有差异,请以英文原文为准。
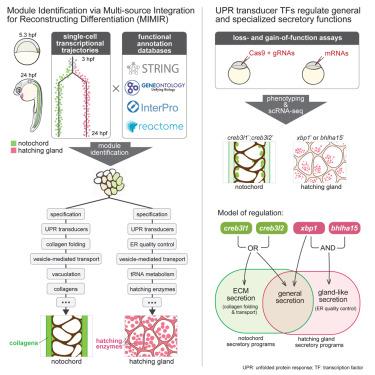
Gene module reconstruction identifies cellular differentiation processes and the regulatory logic of specialized secretion in zebrafish
During differentiation, cells become structurally and functionally specialized, but comprehensive views of the underlying remodeling processes are elusive. Here, we leverage single-cell RNA sequencing (scRNA-seq) developmental trajectories to reconstruct differentiation using two secretory tissues as models—the zebrafish notochord and hatching gland. First, we integrated expression and functional similarities to identify gene modules, revealing dozens of modules representing known and newly associated differentiation processes and their dynamics. Second, we focused on the unfolded protein response (UPR) transducer module to study how general versus cell-type-specific secretory functions are regulated. Profiling loss- and gain-of-function embryos identified that the UPR transcription factors creb3l1, creb3l2, and xbp1 are master regulators of a general secretion program. creb3l1/creb3l2 additionally activate an extracellular matrix secretion program, while xbp1 partners with bhlha15 to activate a gland-like secretion program. Our study presents module identification via multi-source integration for reconstructing differentiation (MIMIR) and illustrates how transcription factors confer general and specialized cellular functions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Developmental cell
生物-发育生物学
CiteScore
18.90
自引率
1.70%
发文量
203
审稿时长
3-6 weeks
期刊介绍:
Developmental Cell, established in 2001, is a comprehensive journal that explores a wide range of topics in cell and developmental biology. Our publication encompasses work across various disciplines within biology, with a particular emphasis on investigating the intersections between cell biology, developmental biology, and other related fields. Our primary objective is to present research conducted through a cell biological perspective, addressing the essential mechanisms governing cell function, cellular interactions, and responses to the environment. Moreover, we focus on understanding the collective behavior of cells, culminating in the formation of tissues, organs, and whole organisms, while also investigating the consequences of any malfunctions in these intricate processes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


