使用基于预训练特征的流式掩码变换器对组蛋白去乙酰化酶抑制剂进行硅学识别。
IF 4.2
3区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
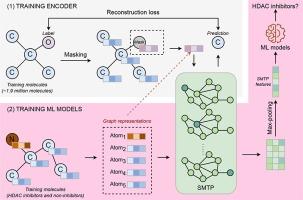
组蛋白去乙酰化酶(HDACs)是一种通过去除组蛋白上的乙酰基来调节基因表达的酶。它们与多种疾病有关,包括神经退行性疾病、心血管疾病、炎症和代谢紊乱,以及肝、肺和肾的纤维化。成功鉴定出强效的 HDAC 抑制剂可能会为治疗这些疾病提供一种前景广阔的方法。除实验技术外,研究人员还引入了几种用于鉴定 HDAC 抑制剂的硅学方法。然而,这些现有的计算机辅助方法在建模阶段存在缺陷,限制了它们的应用。在我们的研究中,我们提出了一种基于简化屏蔽变换器的预训练(SMTP)编码器,可用于生成下游任务的特征。SMTP 编码器的训练过程由基于掩蔽注意力的学习指导,从而增强了模型在编码分子中的通用性。我们利用 SMTP 特征开发了 11 个分类模型,识别了 11 种 HDAC 异构体。我们仅用 190 万个分子对轻量级编码器 SMTP 进行了训练,这比其他已知分子编码器的数量要少,但其判别能力仍然具有竞争力。结果显示,在 11 项分类任务中,使用 SMTP 特征集开发的机器学习模型在 8 项任务中的表现优于使用其他特征集开发的模型。此外,化学多样性分析也证实了编码器在区分两类分子方面的有效性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
In silico identification of Histone Deacetylase inhibitors using Streamlined Masked Transformer-based Pretrained features
Histone Deacetylases (HDACs) are enzymes that regulate gene expression by removing acetyl groups from histones. They are involved in various diseases, including neurodegenerative, cardiovascular, inflammatory, and metabolic disorders, as well as fibrosis in the liver, lungs, and kidneys. Successfully identifying potent HDAC inhibitors may offer a promising approach to treating these diseases. In addition to experimental techniques, researchers have introduced several in silico methods for identifying HDAC inhibitors. However, these existing computer-aided methods have shortcomings in their modeling stages, which limit their applications. In our study, we present a Streamlined Masked Transformer-based Pretrained (SMTP) encoder, which can be used to generate features for downstream tasks. The training process of the SMTP encoder was directed by masked attention-based learning, enhancing the model's generalizability in encoding molecules. The SMTP features were used to develop 11 classification models identifying 11 HDAC isoforms. We trained SMTP, a lightweight encoder, with only 1.9 million molecules, a smaller number than other known molecular encoders, yet its discriminant ability remains competitive. The results revealed that machine learning models developed using the SMTP feature set outperformed those developed using other feature sets in 8 out of 11 classification tasks. Additionally, chemical diversity analysis confirmed the encoder's effectiveness in distinguishing between two classes of molecules.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Methods
生物-生化研究方法
CiteScore
9.80
自引率
2.10%
发文量
222
审稿时长
11.3 weeks
期刊介绍:
Methods focuses on rapidly developing techniques in the experimental biological and medical sciences.
Each topical issue, organized by a guest editor who is an expert in the area covered, consists solely of invited quality articles by specialist authors, many of them reviews. Issues are devoted to specific technical approaches with emphasis on clear detailed descriptions of protocols that allow them to be reproduced easily. The background information provided enables researchers to understand the principles underlying the methods; other helpful sections include comparisons of alternative methods giving the advantages and disadvantages of particular methods, guidance on avoiding potential pitfalls, and suggestions for troubleshooting.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


