在 COVID-19 治疗期间,口咽抗药性基因组保持稳定,而粪便抗药性基因组则向抗药性种类较少的方向转变
IF 4.6
2区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
抗菌药耐药性对全球公共卫生构成严重威胁。COVID-19 大流行凸显了监测抗菌药耐药性基因传播和了解这一过程驱动机制的必要性。在本研究中,我们分析了在医院环境中接受治疗的 COVID-19 患者口咽部和粪便耐药基因组的变化。我们使用了一个包含 4,937 个耐药基因的靶向测序面板来全面描述耐药基因组的特征。我们的研究结果表明,口咽部耐药基因组是均质的,随时间的变化很小。相比之下,粪便样本则分为两种不同的耐药基因型,它们与肠道耐药基因型只有部分关联。约有一半的患者在治疗一周内改变了耐药菌型,其中大多数转变为多样性较低、以ermB为主的耐药菌型2。在80%以上的口咽和粪便样本中都发现了常见的大环内酯类耐药基因,这些基因很可能来自链球菌。我们的研究结果表明,粪便耐药基因组是一个动态系统,可以存在于特定的 "状态 "中,并能从一种状态过渡到另一种状态。迄今为止,这是第一项全面描述口咽部抗药性组及其随时间变化的研究,也是第一项证明粪便抗药性类型随时间变化的研究。本文章由计算机程序翻译,如有差异,请以英文原文为准。
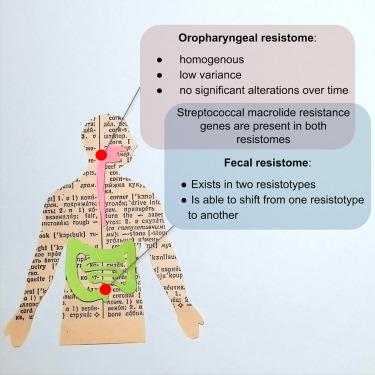
Oropharyngeal resistome remains stable during COVID-19 therapy, while fecal resistome shifts toward a less diverse resistotype
Antimicrobial resistance poses a serious threat to global public health. The COVID-19 pandemic underscored the need to monitor the dissemination of antimicrobial resistance genes and understand the mechanisms driving this process. In this study, we analyzed changes to the oropharyngeal and fecal resistomes of patients with COVID-19 undergoing therapy in a hospital setting. A targeted sequencing panel of 4,937 resistance genes was used to comprehensively characterize resistomes. Our results demonstrated that the oropharyngeal resistome is homogeneous, showing low variability over time. In contrast, fecal samples clustered into two distinct resistotypes that were only partially related to enterotypes. Approximately half of the patients changed their resistotype within a week of therapy, with the majority transitioning to a less diverse and ermB-dominated resistotype 2. Common macrolide resistance genes were identified in over 80% of both oropharyngeal and fecal samples, likely originating from streptococci. Our findings suggest that the fecal resistome is a dynamic system that can exist in certain “states” and is capable of transitioning from one state to another. To date, this is the first study to comprehensively describe the oropharyngeal resistome and its variability over time, and one of the first studies to demonstrate the temporal dynamics of the fecal resistotypes.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

iScience
Multidisciplinary-Multidisciplinary
CiteScore
7.20
自引率
1.70%
发文量
1972
审稿时长
6 weeks
期刊介绍:
Science has many big remaining questions. To address them, we will need to work collaboratively and across disciplines. The goal of iScience is to help fuel that type of interdisciplinary thinking. iScience is a new open-access journal from Cell Press that provides a platform for original research in the life, physical, and earth sciences. The primary criterion for publication in iScience is a significant contribution to a relevant field combined with robust results and underlying methodology. The advances appearing in iScience include both fundamental and applied investigations across this interdisciplinary range of topic areas. To support transparency in scientific investigation, we are happy to consider replication studies and papers that describe negative results.
We know you want your work to be published quickly and to be widely visible within your community and beyond. With the strong international reputation of Cell Press behind it, publication in iScience will help your work garner the attention and recognition it merits. Like all Cell Press journals, iScience prioritizes rapid publication. Our editorial team pays special attention to high-quality author service and to efficient, clear-cut decisions based on the information available within the manuscript. iScience taps into the expertise across Cell Press journals and selected partners to inform our editorial decisions and help publish your science in a timely and seamless way.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


