受到Edwardsiella piscicida挑战的橄榄鲽(Paralichthys olivaceus)的血浆外泌体:表征和 miRNA 分析用于筛选潜在的生物标记物。
IF 4.1
2区 农林科学
Q1 FISHERIES
引用次数: 0
摘要
外泌体从多种类型的细胞中释放出来,这既是正常生理过程的一部分,也是获得性异常过程的一部分。在这项研究中,我们从形态、理化和分子水平研究了致病性Edwardsiella piscicida感染对橄榄鲽(Paralichthys olivaceus)外泌体的影响。采用超速离心法从未感染(PBS-Exo)和实验性感染(Ep-Exo)的橄榄鲽血浆中分离出独特的杯状外泌体。PBS-Exo 的平均粒径、浓度和 zeta 电位分别为 150.9 ± 6.9 nm、5.67 × 1010 颗粒/毫升和 -25.6 ± 1.36 mV,而 Ep-Exo 的平均粒径、浓度和 zeta 电位分别为 138.7 ± 1.9 nm、1.22 × 1011 颗粒/毫升和 -35 ± 1.82 mV。四泛素标记物(CD81、CD9和CD63)的表达证实了橄榄鲽外泌体的存在。在Ep-Exo中发现了差异表达(DE)的已知(9个)和新型(29个)miRNAs(log2折叠变化≥1;p < 0.05),这些miRNAs有可能成为感染的诊断生物标志物。基因本体(Gene Ontology)和京都基因组百科全书(Kyoto Encyclopedia of Genes and Genomes)的通路分析表明,DE miRNA 的预测靶基因高度富集于代谢和免疫作用中。PBS-Exo和Ep-Exo在体外(高达100微克/毫升)和体内(高达400微克/毫升)均无毒性。与载体相比,50 μg/mL的PBS-Exo可诱导Nf-kB(>1.50倍),而100 μg/mL的PBS-Exo可诱导黑头鲦鱼(FHMs)细胞中的Il8、Il10、Nf-kB、P53和Inf(>1.50倍)。这表明 PBS-Exo 中含有适度刺激基因表达的分子。未来,验证Ep-Exo中与DE miRNA相互作用的橄榄鲆免疫反应靶点,对于研究橄榄鲆感染E. piscicida的宿主-病原体相互作用、免疫防御机制和治疗靶点至关重要。本文章由计算机程序翻译,如有差异,请以英文原文为准。
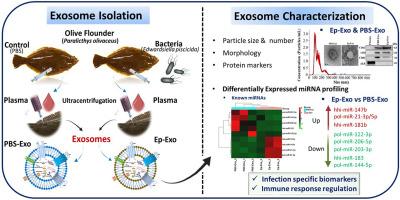
Plasma-derived exosomes of Edwardsiella piscicida challenged olive flounder (Paralichthys olivaceus): Characterization and miRNA profiling for potential biomarkers screening
Exosomes are released from multiple cell types as part of their normal physiology as well as during acquired abnormalities. In this study, we investigated the effect of pathogenic Edwardsiella piscicida infection on olive flounder (Paralichthys olivaceus) exosomes at morphometric, physicochemical, and molecular levels. Unique cup-shaped exosomes were isolated from the plasma of non-infected (PBS-Exo) and E. piscicida experimentally challenged (Ep-Exo) olive flounder using ultracentrifugation. The average particle size, concentration, and zeta potential were 150.9 ± 6.9 nm, 5.67 × 1010 particles/mL, and −25.6 ± 1.36 mV for PBS-Exo while 138.7 ± 1.9 nm, 1.22 × 1011 particles/mL, and −35 ± 1.82 mV for Ep-Exo, respectively. Expression of tetraspanin markers (CD81, CD9, and CD63) confirmed the presence of olive flounder exosomes. Differentially expressed (DE) known (9) and novel (29) miRNAs (log2 fold change ≥1; p < 0.05) were identified in the Ep-Exo that could be potential as diagnostic biomarkers for the infection. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses revealed that the predicted target genes of the DE miRNAs were highly enriched in metabolic and immune roles. Both PBS-Exo and Ep-Exo were non-toxic in vitro (up to 100 μg/mL) and in vivo (up to 400 μg/mL). Compared to the vehicle, PBS-Exo at 50 μg/mL induced Nf-kB (>1.50-fold) while at 100 μg/mL, Il8, Il10, Nf-kB, P53, and Inf were induced (>1.50-fold) in fathead minnow cells (FHMs). This suggests that the PBS-Exo contains molecules that moderately stimulate gene expression. In the future, validating the exact olive flounder immune response target genes that interact with DE miRNAs in Ep-Exo will be crucial for investigating the host-pathogen interactions, immune defense mechanisms, and therapeutic targets for olive flounder against E. piscicida infection.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Fish & shellfish immunology
农林科学-海洋与淡水生物学
CiteScore
7.50
自引率
19.10%
发文量
750
审稿时长
68 days
期刊介绍:
Fish and Shellfish Immunology rapidly publishes high-quality, peer-refereed contributions in the expanding fields of fish and shellfish immunology. It presents studies on the basic mechanisms of both the specific and non-specific defense systems, the cells, tissues, and humoral factors involved, their dependence on environmental and intrinsic factors, response to pathogens, response to vaccination, and applied studies on the development of specific vaccines for use in the aquaculture industry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


