近乎完整的基因组揭示了棉瓜蚜 Aphis gossypii 的种群进化。
IF 3.2
2区 农林科学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
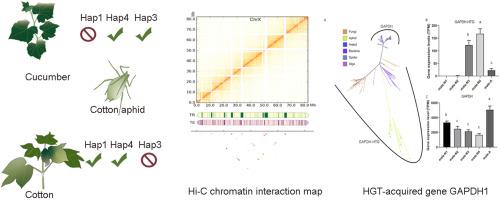
棉瓜蚜 Aphis gossypii Glover 是世界上一种严重的害虫。组间基因组变异可作为分析棉铃蚜适应性的起点。在这项研究中,我们利用长线程 PacBio HiFi 测序和 HiC 搭架技术,对 Hap4 进行了近端粒到端粒的无间隙基因组组装。该装配有两个缺口,总计 321.24 Mb。我们鉴定了五个端粒重复区(GGTTA)n,包括染色体 3' 端的四个重复区,并获得了端粒的新结构信息。由于测序技术的改进,我们还在 Hap4 的基因组组装中发现了超过 55.03 Mb 的重复性 DNA,与 Hap1 和 Hap3 相比,这大大增加了基因组的大小。大部分额外的重复 DNA 位于 X 染色体上,串联重复序列占据了 X 染色体长度的 16.8%。Hap4 组合显示,X 染色体上富含 AT 的 satDNA 阵列(11 个长度超过 100 kb 的 satDNA 阵列)比常染色体上的更多(A1 和 A2 分别含有 3 个和 1 个 satDNA 阵列)。我们检测到了 Hap1、Hap3 和 Hap4 Ap. gossypii 之间的存在-不存在变异、插入和缺失事件,它们对基因表达有显著影响。此外,我们还在所有 Ap. gossypii 菌株中发现了一种雄性特异性的 3-磷酸甘油醛脱氢酶。这一全面的基因组组装为我们深入了解高度重复区域的结构特征提供了宝贵的信息,并允许进行基因组比较分析,从而促进我们对棉铃虫适应性和多样性的了解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
A near-complete genome reveals the population evolution of the cotton-melon aphid Aphis gossypii
The cotton-melon aphid Aphis gossypii Glover is a severe pest worldwide. Interhaplotype genomic variation can be used as a starting point to analyze the adaptability of Ap. gossypii. In this study, we utilized long-read PacBio HiFi sequencing and HiC scaffolding techniques to assemble a near telomere-to-telomere gap-free genome assembly of Hap4. The assembly had two gaps totaling 321.24 Mb. We characterized five telomeric repetitive regions (GGTTA)n, including the four found at the 3′ end of the chromosomes, and obtained new structural information about the telomeres. Due to the improved sequencing technology, we also identified more than 55.03 Mb of repetitive DNA in the genome assembly of Hap4, which contributed significantly to the increase in genome size compared to that of Hap1 and Hap3. Most of the additional repetitive DNA content was located on the X chromosome, and the tandem repeat sequence occupied 16.8% of the X chromosome length. The Hap4 assembly showed that the X chromosome exhibited a greater abundance of AT-rich satDNA arrays (11 satDNA arrays longer than 100 kb) than that observed in the autosomes (A1 and A2 harboured 3 and 1 satDNA arrays). We detected presence-absence variations, insertions, and deletions events between Hap1, Hap3, and Hap4 Ap. gossypii, which had significant effects on gene expression. Additionally, we identified a male-specific glyceraldehyde-3-phosphate dehydrogenase of fungal origin in all strains of Ap. gossypii. This comprehensive genome assembly provides valuable insights into the structural characteristics of highly repetitive regions and allows comparative genomic analyses that facilitate our understanding of Ap. gossypii's adaptation and diversification.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
7.40
自引率
5.30%
发文量
105
审稿时长
40 days
期刊介绍:
This international journal publishes original contributions and mini-reviews in the fields of insect biochemistry and insect molecular biology. Main areas of interest are neurochemistry, hormone and pheromone biochemistry, enzymes and metabolism, hormone action and gene regulation, gene characterization and structure, pharmacology, immunology and cell and tissue culture. Papers on the biochemistry and molecular biology of other groups of arthropods are published if of general interest to the readership. Technique papers will be considered for publication if they significantly advance the field of insect biochemistry and molecular biology in the opinion of the Editors and Editorial Board.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


