铁还原酶氧化酶(FRO)基因的全基因组鉴定和表达分析揭示了它们在非生物和生物胁迫下对铁平衡的关键作用。
IF 6.1
2区 生物学
Q1 PLANT SCIENCES
引用次数: 0
摘要
铁还原酶氧化酶(FRO)基因在植物的铁吸收和平衡中起着关键作用,但在棉花中还没有研究。在这里,我们鉴定并分析了四种棉花(G. hirsutum、G. barbadense、G. arboreum、G. raimondii)中的 65 个 FRO 同源物(21 个 GhFRO、21 个 GbFRO、11 个 GaFRO、12 个 GrFRO)。FRO 在不同物种中表现出保守的铁还原酶活性和保守的结构域;Ferric_reduct (PF01794)、FAD_binding_8 (PF08022) 和 NAD_binding_6 (PF08030)。理化性质和亚细胞定位分析有助于深入了解 FRO 蛋白的功能特征,它们主要定位在质膜上。系统进化分析划分出 11 个群组,显示出保守和不同的进化模式。基因结构分析揭示了不同的外显子内含子组成。染色体定位显示了 A 和 D 基因组的分布,表明了进化的动态性。合成分析揭示了受到纯化选择的旁系和直系基因对。顺式调控元件分析涉及多种调控机制。表达谱分析突显了不同发育阶段、非生物和生物胁迫条件下的动态调控。GhFRO与Ca++依赖性蛋白激酶-10/28-like(CDPKs10/28-like)和金属转运体天然抗性相关巨噬细胞蛋白6(Nramp6)相互作用,调控金属离子转运和铁稳态。三维蛋白质结构预测表明,FRO 蛋白中存在潜在的配体结合位点。此外,用胁迫诱导剂 MeJA、SA、NaCl 和 PEG 处理叶片 1h、2h、4h 和 6h 后,对所选的 8 个 GhFRO 蛋白进行 qRT-PCR 分析,发现它们的表达均显著下调。总之,这项全面的研究深入揭示了棉花中 FRO 基因的多样性、进化、结构、调控和功能,对了解植物铁稳态和胁迫响应具有重要意义。本文章由计算机程序翻译,如有差异,请以英文原文为准。
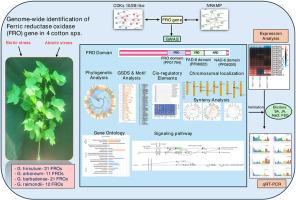
Genome-wide identification and expression analysis of ferric reductase oxidase (FRO) genes in Gossypium spp. reveal their crucial role in iron homeostasis under abiotic and biotic stress
Ferric Reductase Oxidase (FRO) genes are pivotal in iron uptake and homeostasis in plants, yet they are not studied in cotton. Here, we identify and analyze 65 FRO homologs (21 GhFRO, 21 GbFRO, 11 GaFRO, 12 GrFRO) across four Gossypium species (G. hirsutum, G. barbadense, G. arboreum, G. raimondii). FRO exhibit conserved ferric reductase activity and conserved domain structures; Ferric_reduct (PF01794), FAD_binding_8 (PF08022), and NAD_binding_6 (PF08030) across species. Physicochemical properties and subcellular localization analysis provided insights into FRO proteins' functional characteristics, mainly localized to the plasma membrane. Phylogenetic analysis delineates 11 groups, indicating both conserved and divergent evolutionary patterns. Gene structure analysis unveils varying exon-intron compositions. Chromosomal localization shows distribution across A and D genomes, suggesting evolutionary dynamics. Synteny analysis reveals paralogous and orthologous gene pairs subjected to purifying selection. The cis-regulatory elements analysis implicates diverse regulatory mechanisms. Expression profiling highlights dynamic regulation across developmental stages, abiotic and biotic stress conditions. GhFRO interacts with Ca++-dependent protein kinases-10/28-like (CDPKs10/28-like) and metal transporter Natural resistance-associated macrophage protein 6 (Nramp6) to regulate metal ion transport and iron homeostasis. The three-dimensional protein structure prediction suggests potential ligand-binding sites in FRO proteins. Moreover, qRT-PCR analysis of selected eight GhFROs in leaves treated with stress elicitors, MeJA, SA, NaCl, and PEG for 1h, 2h, 4h, and 6h revealed significant downregulation. Overall, this comprehensive study provides insights into FRO gene diversity, evolution, structure, regulation, and function in cotton, with implications for understanding plant iron homeostasis and stress responses.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
11.10
自引率
3.10%
发文量
410
审稿时长
33 days
期刊介绍:
Plant Physiology and Biochemistry publishes original theoretical, experimental and technical contributions in the various fields of plant physiology (biochemistry, physiology, structure, genetics, plant-microbe interactions, etc.) at diverse levels of integration (molecular, subcellular, cellular, organ, whole plant, environmental). Opinions expressed in the journal are the sole responsibility of the authors and publication does not imply the editors'' agreement.
Manuscripts describing molecular-genetic and/or gene expression data that are not integrated with biochemical analysis and/or actual measurements of plant physiological processes are not suitable for PPB. Also "Omics" studies (transcriptomics, proteomics, metabolomics, etc.) reporting descriptive analysis without an element of functional validation assays, will not be considered. Similarly, applied agronomic or phytochemical studies that generate no new, fundamental insights in plant physiological and/or biochemical processes are not suitable for publication in PPB.
Plant Physiology and Biochemistry publishes several types of articles: Reviews, Papers and Short Papers. Articles for Reviews are either invited by the editor or proposed by the authors for the editor''s prior agreement. Reviews should not exceed 40 typewritten pages and Short Papers no more than approximately 8 typewritten pages. The fundamental character of Plant Physiology and Biochemistry remains that of a journal for original results.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


