单分子状态将转录因子结合与基因表达联系起来
IF 50.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
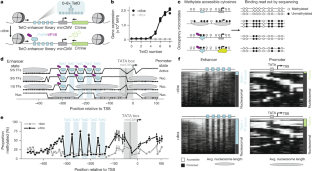
多种转录因子(TF)与基因组增强子的结合推动了哺乳动物细胞中基因的表达1。然而,将增强子序列与 TF 结合、启动子状态和转录水平联系起来的分子细节仍不清楚。在这里,我们采用单分子足迹法2,3测量了TF、核糖体和其他调控蛋白在具有不同数量TF结合位点的工程增强子-启动子构建体上的同时占据情况,这些位点既包括合成TF,也包括参与I型干扰素反应的内源性TF。虽然无核糖体 DNA 上的 TF 结合事件是独立的,但激活结构域招募的辅助因子会破坏核糖体的稳定性,从而推动观察到的 TF 结合合作性。平均 TF 占有率线性地决定了启动子的活性,我们将 TF 强度分解为可分离的结合和激活项。最后,我们建立了热力学和动力学模型,可定量预测增强子结合微观状态和基因表达动态。这项工作为定量分析基因表达的不同贡献者提供了一个模板,包括 TF 激活域、浓度、结合亲和力、结合位点配置和染色质调节因子的招募。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Single-molecule states link transcription factor binding to gene expression
The binding of multiple transcription factors (TFs) to genomic enhancers drives gene expression in mammalian cells1. However, the molecular details that link enhancer sequence to TF binding, promoter state and transcription levels remain unclear. Here we applied single-molecule footprinting2,3 to measure the simultaneous occupancy of TFs, nucleosomes and other regulatory proteins on engineered enhancer–promoter constructs with variable numbers of TF binding sites for both a synthetic TF and an endogenous TF involved in the type I interferon response. Although TF binding events on nucleosome-free DNA are independent, activation domains recruit cofactors that destabilize nucleosomes, driving observed TF binding cooperativity. Average TF occupancy linearly determines promoter activity, and we decompose TF strength into separable binding and activation terms. Finally, we develop thermodynamic and kinetic models that quantitatively predict both the enhancer binding microstates and gene expression dynamics. This work provides a template for the quantitative dissection of distinct contributors to gene expression, including TF activation domains, concentration, binding affinity, binding site configuration and recruitment of chromatin regulators. A study uses single-molecule footprinting to measure protein occupancy at regulatory elements on individual molecules in human cells and describes how different properties of transcription factor binding contribute to gene expression.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


