将天然共生菌和病原体纳入临床前小鼠模型
IF 60.9
1区 医学
Q1 IMMUNOLOGY
引用次数: 0
摘要
以实验小鼠为研究模型,已经在哺乳动物生理学的许多方面取得了重大发现。这些研究得益于小鼠在遗传学上的可操作性和近亲繁殖、大量可用的免疫试剂以及环境可控的高通量设施的建立。这些设施通常包括一些屏障,以保持小鼠群落中没有病原体,而且小鼠的频繁再繁殖严重限制了它们的共生菌群。由于人类与微生物共同进化并接触各种病原体,越来越多的研究人员认为,在小鼠在自然界中会遇到的微生物群和病原体的背景下研究小鼠,可以改进临床前疾病研究。在此,我们将从一个角度来探讨如何将这些不同的方法结合起来并加以整合,以改进现有的小鼠模型,从而提高我们对疾病机理的认识,并为人类开发出新的疗法。我们还提出,与 "脏小鼠 "等现有术语相比,"天然微生物群小鼠 "更适合描述这些模型。本文章由计算机程序翻译,如有差异,请以英文原文为准。
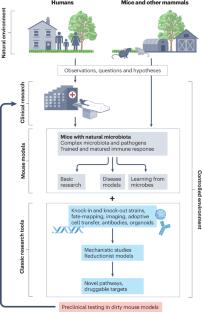
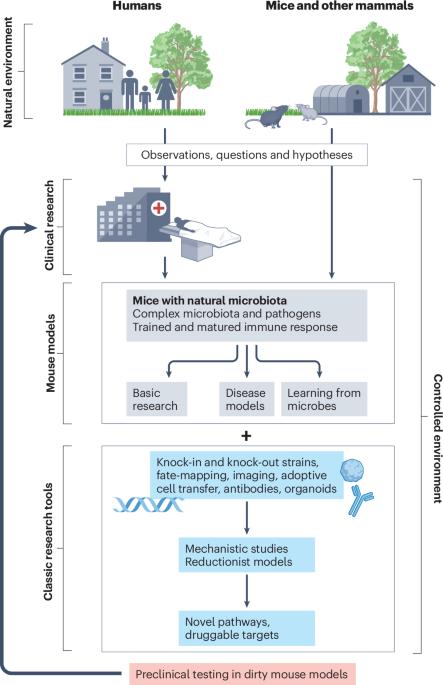
Integrating natural commensals and pathogens into preclinical mouse models
Fundamental discoveries in many aspects of mammalian physiology have been made using laboratory mice as research models. These studies have been facilitated by the genetic tractability and inbreeding of such mice, the large set of immunological reagents that are available, and the establishment of environmentally controlled, high-throughput facilities. Such facilities typically include barriers to keep the mouse colonies free of pathogens and the frequent re-derivation of the mice severely limits their commensal flora. Because humans have co-evolved with microorganisms and are exposed to a variety of pathogens, a growing community of researchers posits that preclinical disease research can be improved by studying mice in the context of the microbiota and pathogens that they would encounter in the natural world. Here, we provide a perspective of how these different approaches can be combined and integrated to improve existing mouse models to enhance our understanding of disease mechanisms and develop new therapies for humans. We also propose that the term ‘mice with natural microbiota’ is more appropriate for describing these models than existing terms such as ‘dirty mice’. There is emerging evidence that mice with a history of microbial exposures can better model the human immune system than laboratory mice maintained in pathogen-free conditions. In this Perspective, Rehermann and colleagues summarize different approaches that have been used to incorporate microbiota and pathogen exposures into laboratory mouse models. They suggest that the term ‘mice with natural microbiota’ should be used instead of ‘dirty mice’ to describe these systems in the future.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Reviews Immunology
医学-免疫学
CiteScore
93.40
自引率
0.40%
发文量
131
审稿时长
6-12 weeks
期刊介绍:
Nature Reviews Immunology is a journal that provides comprehensive coverage of all areas of immunology, including fundamental mechanisms and applied aspects. It has two international standard serial numbers (ISSN): 1474-1733 for print and 1474-1741 for online. In addition to review articles, the journal also features recent developments and new primary papers in the field, as well as reflections on influential people, papers, and events in the development of immunology. The subjects covered by Nature Reviews Immunology include allergy and asthma, autoimmunity, antigen processing and presentation, apoptosis and cell death, chemokines and chemokine receptors, cytokines and cytokine receptors, development and function of cells of the immune system, haematopoiesis, infection and immunity, immunotherapy, innate immunity, mucosal immunology and the microbiota, regulation of the immune response, signalling in the immune system, transplantation, tumour immunology and immunotherapy, and vaccine development.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


