融合多模态信息的分子表征预训练图转换器
IF 14.7
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
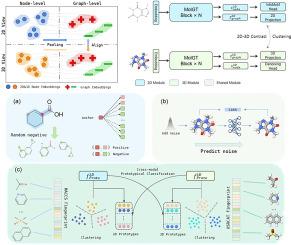
分子表征学习(MRL)在药物发现和生命科学等某些应用中至关重要。尽管分子表征学习在多视角和多模态学习方面取得了进展,但现有模型仅探索了有限的视角范围,而分子表征学习中不同视角和模态的融合仍未得到充分探索。此外,由于计算成本较高,获取分子的几何构象在许多任务中并不可行。为 MRL 设计一个通用的获取模型是值得的,但也是具有挑战性的。本文提出了一种融合节点和图形视图以及分子二维拓扑和三维几何模式的新型图变换器预训练框架,称为 MolGT。这种 MolGT 模型集成了节点级和图级的二维拓扑和三维几何预训练任务,利用定制的模态共享图变换器,在参数效率和跨模态知识共享方面具有多功能性。此外,MolGT 还能利用二维拓扑和三维几何模态之间的对比学习,生成隐式三维几何图形。我们提供了大量实验和深入分析,验证了 MolGT 能够:(1)确实利用多视角和多模态信息准确地表示分子;(2)使用二维分子推断出几乎相同的结果,而无需进行生成构象的昂贵计算。代码可在 GitHub11https://github.com/robbenplus/MolGT 上获取。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Pretraining graph transformer for molecular representation with fusion of multimodal information
Molecular representation learning (MRL) is essential in certain applications including drug discovery and life science. Despite advancements in multiview and multimodal learning in MRL, existing models have explored only a limited range of perspectives, and the fusion of different views and modalities in MRL remains underexplored. Besides, obtaining the geometric conformer of molecules is not feasible in many tasks due to the high computational cost. Designing a general-purpose pertaining model for MRL is worthwhile yet challenging. This paper proposes a novel graph Transformer pretraining framework with fusion of node and graph views, along with the 2D topology and 3D geometry modalities of molecules, called MolGT. This MolGT model integrates node-level and graph-level pretext tasks on 2D topology and 3D geometry, leveraging a customized modality-shared graph Transformer that has versatility regarding parameter efficiency and knowledge sharing across modalities. Moreover, MolGT can produce implicit 3D geometry by leveraging contrastive learning between 2D topological and 3D geometric modalities. We provide extensive experiments and in-depth analyses, verifying that MolGT can (1) indeed leverage multiview and multimodal information to represent molecules accurately, and (2) infer nearly identical results using 2D molecules without requiring the expensive computation of generating conformers. Code is available on GitHub1.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Information Fusion
工程技术-计算机:理论方法
CiteScore
33.20
自引率
4.30%
发文量
161
审稿时长
7.9 months
期刊介绍:
Information Fusion serves as a central platform for showcasing advancements in multi-sensor, multi-source, multi-process information fusion, fostering collaboration among diverse disciplines driving its progress. It is the leading outlet for sharing research and development in this field, focusing on architectures, algorithms, and applications. Papers dealing with fundamental theoretical analyses as well as those demonstrating their application to real-world problems will be welcome.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


