利用QSAR、ADMET、分子对接、MM-GBSA和分子动力学模拟方法,合理设计一些1,3,4三取代吡唑-噻唑衍生物作为MtInhA抑制剂
IF 3.8
Q2 CHEMISTRY, PHYSICAL
引用次数: 0
摘要
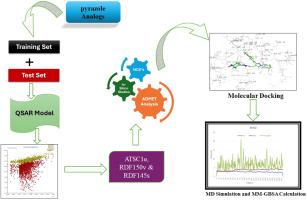
利用计算方法预测了 1,3,4 三取代吡唑衍生物作为 MtInhA 抑制剂的潜在功效和特异性,这将有助于结核病治疗的合理药物设计。采用 QSARINS 软件研究了化合物抑制 MtInhA 的能力。生成的 QSAR 模型确定了三个具有显著相关性和令人印象深刻的统计值的值得注意的描述因子:相关系数(R2)=0.8789;交叉验证剔除一个相关系数(Q2LOO)=0.8402;交叉验证剔除多个相关系数(Q2LMO)=0.7321;交叉验证相关系数(CCCtr)=0.9355;CCCext=0.888。QSAR 模型生成的描述符包括以桑德森电负性(ATSC1e)加权的居中布罗托-莫罗自相关性、以相对范德华体积(RDF150v)加权的 15.0 和 2.0 Å 原子间距离径向分布函数、以相对 I 态(RDF145s)加权的径向分布函数 - 145/。利用这些方法,建立了三个描述符模型,并对设计的化合物进行了 MtInhA 抑制活性评估。此外,还使用 Schrodinger 软件进行了 ADMET 预测和分子对接研究。ADMET 预测用于评估药物的相似性,分子对接用于确定设计化合物与目标蛋白质的相互作用。对接研究结束后,对化合物进行了 MM-GBSA 计算和 MD 模拟。在设计的化合物中,AP2 与 MtInhA 酶的结合亲和力最强。这项工作的结果有助于了解 1,3,4 三取代吡唑衍生物与 MtInhA 蛋白之间的关键相互作用,这可能是开发新的抗结核先导化合物所必需的。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Rational design of some 1,3,4 trisubstituted pyrazole-thiazole derivatives to serve as MtInhA inhibitors using QSAR, ADMET, molecular docking, MM-GBSA, and molecular dynamics simulations approach
Using computational approaches, the potential efficacy and specificity of 1,3,4 trisubstituted pyrazole derivatives as MtInhA inhibitors which will aid in rational drug design for tubercular therapy were forecasted. QSARINS software was used to investigate the ability of compound to inhibit MtInhA. Three noteworthy descriptors with significant correlations and impressive statistical values were identified by the produced QSAR model: Correlation of coefficient(R2)= 0.8789, Cross-validation leave one out correlation coefficient (Q2LOO)= 0.8402, Cross-validation leave-many-out correlation coefficient(Q2LMO)=0.7321, Concordance Correlation Coefficient for cross-validation(CCCtr)=0.9355, CCCext =0.888. The descriptor generated by the QSAR model includes Centered Broto-Moreau autocorrelation weighted by Sanderson electronegativities (ATSC1e), Radial distribution functions at 15.0 and 2.0 Å inter-atomic distances weighted by relative van der Waals volumes (RDF150v), Radial distribution function – 145/weighted by relative I-state (RDF145s). Using these, three descriptor model was developed and the designed compounds were evaluated for their MtInhA inhibitory activity. Further, ADMET prediction and Molecular docking studies were carried out using Schrodinger's Software. ADMET prediction were used to evaluate drug likeliness and molecular docking was used to determine the interactions of designed compounds with the target protein. After the docking studies, the compounds were subjected for MM-GBSA calculations and MD simulation. Among the designed compounds, AP2 had the strongest binding affinity towards the MtInhA enzyme. The result of this work helps to understand the key interactions between 1,3,4 trisubstituted pyrazole derivatives and MtInhA protein that may be necessary to develop new lead compounds against tuberculosis.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Chemical Physics Impact
Materials Science-Materials Science (miscellaneous)
CiteScore
2.60
自引率
0.00%
发文量
65
审稿时长
46 days
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


