通过预筛选类似 ARG 的读数快速识别抗生素耐药基因宿主
IF 14.3
1区 环境科学与生态学
Q1 ENVIRONMENTAL SCIENCES
引用次数: 0
摘要
有效的风险评估和环境抗生素耐药性控制取决于有关抗生素耐药性基因(ARGs)及其微生物宿主的全面信息。测序技术和生物信息学的进步使得利用元基因组组装的等位基因和基因组识别 ARG 宿主成为可能。然而,这些方法往往会造成信息丢失,而且需要大量的计算资源。在这里,我们介绍了一种生物信息学策略,通过直接从元基因组数据集中预选类似 ARG 的读数(ALR)来鉴定 ARG 宿主。这种基于 ALR 的方法有几个优点:(1)它能在复杂环境中更准确地检测低丰度 ARG 宿主;(2)它在 ARG 的丰度和宿主之间建立了直接的关系;(3)与依赖于组装的等位基因组和基因组的策略相比,它减少了大约 44-96% 的计算时间。我们将基于 ALR 的策略与两种传统方法一起用于研究典型的人类影响环境。所有方法的结果都是一致的,揭示出 ARGs 主要由伽马蛋白菌和芽孢杆菌携带,它们的分布模式可能表明废水排放对沿岸抗性组的影响。我们的策略能快速准确地鉴定抗生素耐药菌,为高通量监测环境抗生素耐药性提供了宝贵的见解。这项研究进一步拓展了我们对未来 ARG 相关风险管理的认识。本文章由计算机程序翻译,如有差异,请以英文原文为准。
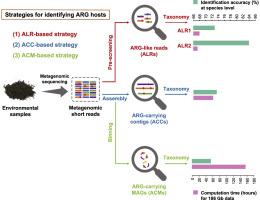
Rapid identification of antibiotic resistance gene hosts by prescreening ARG-like reads
Effective risk assessment and control of environmental antibiotic resistance depend on comprehensive information about antibiotic resistance genes (ARGs) and their microbial hosts. Advances in sequencing technologies and bioinformatics have enabled the identification of ARG hosts using metagenome-assembled contigs and genomes. However, these approaches often suffer from information loss and require extensive computational resources. Here we introduce a bioinformatic strategy that identifies ARG hosts by prescreening ARG-like reads (ALRs) directly from total metagenomic datasets. This ALR-based method offers several advantages: (1) it enables the detection of low-abundance ARG hosts with higher accuracy in complex environments; (2) it establishes a direct relationship between the abundance of ARGs and their hosts; and (3) it reduces computation time by approximately 44–96% compared to strategies relying on assembled contigs and genomes. We applied our ALR-based strategy alongside two traditional methods to investigate a typical human-impacted environment. The results were consistent across all methods, revealing that ARGs are predominantly carried by Gammaproteobacteria and Bacilli, and their distribution patterns may indicate the impact of wastewater discharge on coastal resistome. Our strategy provides rapid and accurate identification of antibiotic-resistant bacteria, offering valuable insights for the high-throughput surveillance of environmental antibiotic resistance. This study further expands our knowledge of ARG-related risk management in future.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Environmental Science and Ecotechnology
Multiple-
CiteScore
20.40
自引率
6.30%
发文量
11
审稿时长
18 days
期刊介绍:
Environmental Science & Ecotechnology (ESE) is an international, open-access journal publishing original research in environmental science, engineering, ecotechnology, and related fields. Authors publishing in ESE can immediately, permanently, and freely share their work. They have license options and retain copyright. Published by Elsevier, ESE is co-organized by the Chinese Society for Environmental Sciences, Harbin Institute of Technology, and the Chinese Research Academy of Environmental Sciences, under the supervision of the China Association for Science and Technology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


