基于转录组的败血症风险模型可进行预后预测和药物重新定位
IF 4.6
2区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
由于宿主反应的异质性,脓毒症的治疗面临巨大挑战。我们的目标是开发一种基于转录组特征的早期脓毒症分层风险模型。该模型在脓毒症诊断(AUC = 0.99-1.00)和预后(AUC = 0.61-0.70)方面表现优异,优于 Mars 和 SRS 内型。此外,该模型还揭示了高危患者的免疫抑制状态和低危患者对氢化可的松的不良反应。单细胞转录组分析进一步阐明了四种基因在不同免疫细胞类型中的表达模式和影响,说明了该模型所反映的综合宿主反应。根据风险亚组不同的转录特征,我们确定了芬瑞替尼和美洛昔康作为治疗药物,这两种药物能显著提高脓毒症小鼠模型的存活率。我们的研究引入了一种风险模型,可优化脓毒症的风险分层和药物再利用,从而提供一种全面的管理方法。本文章由计算机程序翻译,如有差异,请以英文原文为准。
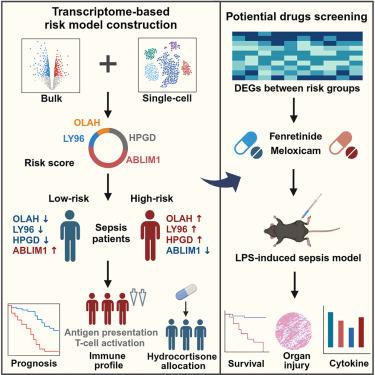
A transcriptome-based risk model in sepsis enables prognostic prediction and drug repositioning
Septic management presented a tremendous challenge due to heterogeneous host responses. We aimed to develop a risk model for early septic stratification based on transcriptomic signature. Here, we combined genes OLAH, LY96, HPGD, and ABLIM1 into a prognostic risk score model, which demonstrated exceptional performance in septic diagnosis (AUC = 0.99–1.00) and prognosis (AUC = 0.61–0.70), outperforming that of Mars and SRS endotypes. Also, the model unveiled immunosuppressive status in high-risk patients and a poor response to hydrocortisone in low-risk individuals. Single-cell transcriptome analysis further elucidated expression patterns and effects of the four genes across immune cell types, illustrating integrated host responses reflected by this model. Upon distinct transcriptional profiles of risk subgroups, we identified fenretinide and meloxicam as therapeutic agents, which significantly improved survival in septic mice models. Our study introduced a risk model that optimized risk stratification and drug repurposing of sepsis, thereby offering a comprehensive management approach.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

iScience
Multidisciplinary-Multidisciplinary
CiteScore
7.20
自引率
1.70%
发文量
1972
审稿时长
6 weeks
期刊介绍:
Science has many big remaining questions. To address them, we will need to work collaboratively and across disciplines. The goal of iScience is to help fuel that type of interdisciplinary thinking. iScience is a new open-access journal from Cell Press that provides a platform for original research in the life, physical, and earth sciences. The primary criterion for publication in iScience is a significant contribution to a relevant field combined with robust results and underlying methodology. The advances appearing in iScience include both fundamental and applied investigations across this interdisciplinary range of topic areas. To support transparency in scientific investigation, we are happy to consider replication studies and papers that describe negative results.
We know you want your work to be published quickly and to be widely visible within your community and beyond. With the strong international reputation of Cell Press behind it, publication in iScience will help your work garner the attention and recognition it merits. Like all Cell Press journals, iScience prioritizes rapid publication. Our editorial team pays special attention to high-quality author service and to efficient, clear-cut decisions based on the information available within the manuscript. iScience taps into the expertise across Cell Press journals and selected partners to inform our editorial decisions and help publish your science in a timely and seamless way.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


