从采样到模拟:系统病理生理学建模中的单细胞多组学
IF 4.6
2区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
随着单细胞全息数据取样和采集方法以前所未有的速度积累,人们开发出了各种数据分析管道,用于推断细胞类型、细胞状态及其分布、状态转换、状态轨迹和状态相互作用。这带来了一个新的机遇,即可以利用单细胞组学数据生成高分辨率、高保真的计算模型。在本综述中,我们将讨论如何利用单细胞全息数据建立计算模型,以模拟各种尺度的生物系统。我们提出,单细胞数据可与生理信息相结合,生成器官特异性模型,然后将这些模型组合起来,生成多器官系统病理生理学模型。最后,我们讨论了如何将通用的多器官模型提升到患者特异性水平,从而允许在临床环境中使用这些模型。本文章由计算机程序翻译,如有差异,请以英文原文为准。
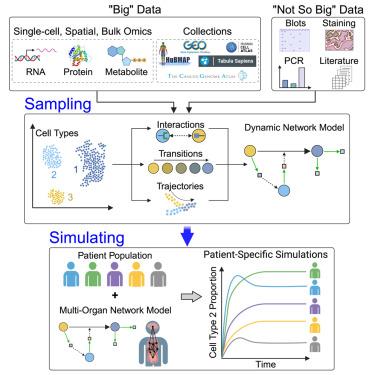
From sampling to simulating: Single-cell multiomics in systems pathophysiological modeling
As single-cell omics data sampling and acquisition methods have accumulated at an unprecedented rate, various data analysis pipelines have been developed for the inference of cell types, cell states and their distribution, state transitions, state trajectories, and state interactions. This presents a new opportunity in which single-cell omics data can be utilized to generate high-resolution, high-fidelity computational models. In this review, we discuss how single-cell omics data can be used to build computational models to simulate biological systems at various scales. We propose that single-cell data can be integrated with physiological information to generate organ-specific models, which can then be assembled to generate multi-organ systems pathophysiological models. Finally, we discuss how generic multi-organ models can be brought to the patient-specific level thus permitting their use in the clinical setting.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

iScience
Multidisciplinary-Multidisciplinary
CiteScore
7.20
自引率
1.70%
发文量
1972
审稿时长
6 weeks
期刊介绍:
Science has many big remaining questions. To address them, we will need to work collaboratively and across disciplines. The goal of iScience is to help fuel that type of interdisciplinary thinking. iScience is a new open-access journal from Cell Press that provides a platform for original research in the life, physical, and earth sciences. The primary criterion for publication in iScience is a significant contribution to a relevant field combined with robust results and underlying methodology. The advances appearing in iScience include both fundamental and applied investigations across this interdisciplinary range of topic areas. To support transparency in scientific investigation, we are happy to consider replication studies and papers that describe negative results.
We know you want your work to be published quickly and to be widely visible within your community and beyond. With the strong international reputation of Cell Press behind it, publication in iScience will help your work garner the attention and recognition it merits. Like all Cell Press journals, iScience prioritizes rapid publication. Our editorial team pays special attention to high-quality author service and to efficient, clear-cut decisions based on the information available within the manuscript. iScience taps into the expertise across Cell Press journals and selected partners to inform our editorial decisions and help publish your science in a timely and seamless way.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


