对医疗废弃物进行浅层射枪测序发现具有广谱抗生素基因的食塑料细菌
IF 7.3
2区 环境科学与生态学
Q1 ENVIRONMENTAL SCIENCES
引用次数: 0
摘要
抗菌素耐药性和塑料污染的危机正在医疗保健领域迅速蔓延,给全球健康带来了复杂的挑战。本研究调查了医疗废物中的微生物种群,以了解抗菌药耐药性的程度以及细菌降解塑料的潜力。我们采用扩增子和浅层枪式测序技术进行元基因组分析,全面了解了微生物群的分类多样性和功能能力。我们采用全长 16S rRNA 测序法对医疗废物样本中的存活细菌进行了分析,结果表明细菌群落的多样性以固着菌门和变形菌门为主。值得注意的是,样本中检测到了奇异变形杆菌 VFC3/3 和假单胞菌 VFA2/3,而嗜麦芽气单胞菌 VFV3/2 则是主要物种,这对医院获得性感染的传播和抗菌药耐药性产生了影响。抗生素药敏试验发现,耐多药菌株具有抗菌基因,包括广谱抗生素碳青霉烯,这突出表明了改善废物管理和感染控制措施的迫切需要。值得注意的是,我们发现了与塑料分解有关的基因,这些基因编码酯酶、解聚酶和氧化还原酶类的酶。这表明,在医疗废物中发现的特定细菌可能能够减少生物和医疗废物造成的塑料污染。这些信息有助于制定应对环境污染和抗生素耐药性等综合问题的策略。这项研究强调了监测医院废物中微生物群落以影响废物管理程序和公共卫生政策的重要性。研究结果突出表明,有必要采用多学科方法来降低与抗生素耐药性和塑料垃圾相关的风险,尤其是在两者交叉最为严重的医院环境中。本文章由计算机程序翻译,如有差异,请以英文原文为准。
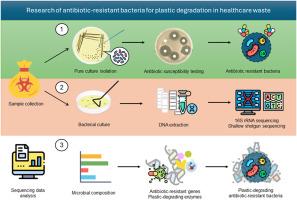

Shallow shotgun sequencing of healthcare waste reveals plastic-eating bacteria with broad-spectrum antibiotic resistance genes
The burgeoning crises of antimicrobial resistance and plastic pollution are converging in healthcare settings, presenting a complex challenge to global health. This study investigates the microbial populations in healthcare waste to understand the extent of antimicrobial resistance and the potential for plastic degradation by bacteria. Our metagenomic analysis, using both amplicon and shallow shotgun sequencing, provided a comprehensive view of the taxonomic diversity and functional capacity of the microbial consortia. The viable bacteria in healthcare waste samples were analyzed employing full-length 16S rRNA sequencing, revealing a diverse bacterial community dominated by Firmicutes and Proteobacteria phyla. Notably, Proteus mirabilis VFC3/3 and Pseudomonas sp. VFA2/3 were detected, while Stenotrophomonas maltophilia VFV3/2 surfaced as the predominant species, holding implications for the spread of hospital-acquired infections and antimicrobial resistance. Antibiotic susceptibility testing identified multidrug-resistant strains conferring antimicrobial genes, including the broad-spectrum antibiotic carbapenem, underscoring the critical need for improved waste management and infection control measures. Remarkably, we found genes linked to the breakdown of plastic that encoded for enzymes of the esterase, depolymerase, and oxidoreductase classes. This suggests that specific bacteria found in medical waste may be able to reduce the amount of plastic pollution that comes from biological and medical waste. The information is helpful in formulating strategies to counter the combined problems of environmental pollution and antibiotic resistance. This study emphasises the importance of monitoring microbial communities in hospital waste in order to influence waste management procedures and public health policy. The findings highlight the need for a multidisciplinary approach to mitigate the risks associated with antimicrobial resistance and plastic waste, especially in hospital settings where they intersect most acutely.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Environmental Pollution
环境科学-环境科学
CiteScore
16.00
自引率
6.70%
发文量
2082
审稿时长
2.9 months
期刊介绍:
Environmental Pollution is an international peer-reviewed journal that publishes high-quality research papers and review articles covering all aspects of environmental pollution and its impacts on ecosystems and human health.
Subject areas include, but are not limited to:
• Sources and occurrences of pollutants that are clearly defined and measured in environmental compartments, food and food-related items, and human bodies;
• Interlinks between contaminant exposure and biological, ecological, and human health effects, including those of climate change;
• Contaminants of emerging concerns (including but not limited to antibiotic resistant microorganisms or genes, microplastics/nanoplastics, electronic wastes, light, and noise) and/or their biological, ecological, or human health effects;
• Laboratory and field studies on the remediation/mitigation of environmental pollution via new techniques and with clear links to biological, ecological, or human health effects;
• Modeling of pollution processes, patterns, or trends that is of clear environmental and/or human health interest;
• New techniques that measure and examine environmental occurrences, transport, behavior, and effects of pollutants within the environment or the laboratory, provided that they can be clearly used to address problems within regional or global environmental compartments.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


