利用慕尼黑亥姆霍兹基因型估算服务器实现 GDPR 合规性
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
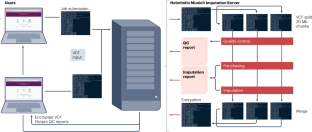
基因组学有可能彻底改变医疗保健,使个性化疾病管理(包括精准预防)成为可能。全基因组关联研究(GWAS)有助于对复杂的人类疾病产生新的生物学见解1。通过荟萃分析增加样本量可以提高全基因组关联研究的能力,而荟萃分析需要对某些研究中可能未分型的基因型进行估算和分析。估算依赖于全基因组测序个体的分期单倍型参考面板2。由于数据访问和安全性、数据集大小以及实现估算所需的计算资源规模等原因,这些数据无法与需要估算其 GWAS 数据的研究人员共享。因此,人们开发了估算服务器来提供解决方案:研究人员将他们的基因分型数据集上传到托管参考面板和估算机器的估算服务器上,在那里对数据进行估算,然后下载回研究人员各自的本地计算环境。目前有许多为全球研究人员服务的估算服务器,包括两个位于美国的服务器(密歇根大学 https://imputationserver.sph.umich.edu/index.html 和 TOPMed,https://imputation.biodatacatalyst.nhlbi.nih.gov/)、一个位于英国的服务器(惠康桑格研究所 https://imputation.sanger.ac.uk/?about=1)和一个位于德国基尔大学的服务器(https://hybridcomputing.ikmb.uni-kiel.de)。在此,我们开发了一个基于欧盟(EU)的估算服务器,为位于德国慕尼黑的整个社区服务(https://imputationserver.helmholtz-munich.de/),以帮助用户遵守《一般数据保护条例》(GDPR)的要求。之所以需要基于欧盟的估算服务器,是因为欧盟的全面数据隐私法(GDPR law3)施加了限制。根据 GDPR,基因数据被视为特殊类别的个人数据,因此必须遵守严格的数据共享规则和保障措施4。将基因型数据上传到不在欧盟境内或不在适足性协议范围内的估算服务器构成违反 GDPR 的行为,除非相关研究的知情同意书中有明确规定。在此,我们介绍亥姆霍兹慕尼黑估算服务器,该服务器旨在以符合 GDPR 的方式为欧盟以及全球的研究人员提供免费的基因型估算服务。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Toward GDPR compliance with the Helmholtz Munich genotype imputation server
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


