利用噬菌体 T7 检测大肠杆菌并分析激发-发射矩阵荧光光谱。
IF 2.1
4区 农林科学
Q3 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
摘要
传统的检测方法需要分离和富集细菌,然后进行分子、生化或培养分析。为了解决传统方法的一些局限性,本研究开发了一种机器学习(ML)方法,用于分析基于噬菌体 T7 和大肠杆菌相互作用生成的激发-发射矩阵(EEM)荧光数据,以便在存在新鲜农产品匀浆的情况下原位检测活细菌。我们根据细菌及其与 T7 噬菌体相互作用生成的三维 EEM 数据,使用各种 ML 算法训练分类模型。包括线性支持向量分类器和随机森林在内的这些 ML 算法在 6 小时内检测 102 CFU/ml 浓度的大肠杆菌方面表现出很高的准确性(>0.85)。此外,这些 ML 模型还能区分不同的大肠杆菌浓度水平。例如,高斯过程模型检测不同浓度水平的活大肠杆菌的准确率达到 92%。应用这些 ML 方法检测菠菜匀浆中的大肠杆菌时,线性-SVC 模型的准确率为 89%。此外,还采用了特征选择技术来降低数据的维度,结果表明,只需要六个特征就能对含有 102 CFU/ml 大肠杆菌的菠菜匀浆样本达到分类准确率(大于 0.85)。这些发现凸显了这种新型细菌检测方法的潜力,为食品安全和环境监测应用提供了快速、特异和高效的解决方案。本文章由计算机程序翻译,如有差异,请以英文原文为准。
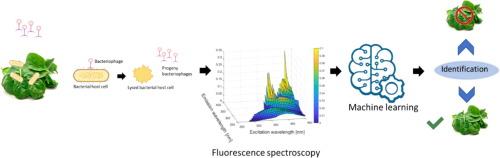
Detection of Escherichia coli Using Bacteriophage T7 and Analysis of Excitation‑Emission Matrix Fluorescence Spectroscopy
Conventional detection methods require the isolation and enrichment of bacteria, followed by molecular, biochemical, or culture-based analysis. To address some of the limitations of conventional methods, this study develops a machine learning (ML) approach to analyze the excitation-emission matrix (EEM) fluorescence data generated based on bacteriophage T7 and Escherichia coli interactions for in-situ detection of live bacteria in the presence of fresh produce homogenate. We trained classification models using various ML algorithms based on the 3-D EEM data generated with bacteria and their interactions with a T7 phage. These ML algorithms, including linear Support Vector Classifier (SVC) and Random Forest (RF), demonstrate high accuracy (>0.85) for detecting E. coli at 102 CFU/ml concentration within 6 h. Additionally, these ML models can differentiate among different E. coli concentration levels. For example, the Gaussian Process model achieved an accuracy of 92% in detecting different concentration levels of live E. coli. Application of these ML methods to detect E. coli in spinach homogenate yielded an accuracy of 89% using the linear-SVC model. Furthermore, feature selection techniques were employed to reduce the dimensionality of the data, revealing that only six features were necessary for achieving classification accuracy (>0.85) of spinach homogenate samples containing 102 CFU/ml of E. coli. These findings highlight the potential of this novel bacterial detection methodology, offering rapid, specific, and efficient solutions for applications in food safety and environmental monitoring.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of food protection
工程技术-生物工程与应用微生物
CiteScore
4.20
自引率
5.00%
发文量
296
审稿时长
2.5 months
期刊介绍:
The Journal of Food Protection® (JFP) is an international, monthly scientific journal in the English language published by the International Association for Food Protection (IAFP). JFP publishes research and review articles on all aspects of food protection and safety. Major emphases of JFP are placed on studies dealing with:
Tracking, detecting (including traditional, molecular, and real-time), inactivating, and controlling food-related hazards, including microorganisms (including antibiotic resistance), microbial (mycotoxins, seafood toxins) and non-microbial toxins (heavy metals, pesticides, veterinary drug residues, migrants from food packaging, and processing contaminants), allergens and pests (insects, rodents) in human food, pet food and animal feed throughout the food chain;
Microbiological food quality and traditional/novel methods to assay microbiological food quality;
Prevention of food-related hazards and food spoilage through food preservatives and thermal/non-thermal processes, including process validation;
Food fermentations and food-related probiotics;
Safe food handling practices during pre-harvest, harvest, post-harvest, distribution and consumption, including food safety education for retailers, foodservice, and consumers;
Risk assessments for food-related hazards;
Economic impact of food-related hazards, foodborne illness, food loss, food spoilage, and adulterated foods;
Food fraud, food authentication, food defense, and foodborne disease outbreak investigations.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


