一种基于 DNA 酶介导的信号放大的简单、无酶的 p53 敏感分析方法。
IF 2.6
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
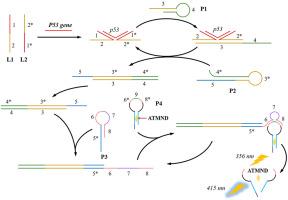
目前迫切需要一种简单而又极其精确的生物传感器来分析肿瘤的发生。在此,我们提出了一种新型的荧光和无酶方法,用于检测 p53 基因级联近接连接介导的催化发夹组装和 DNA 酶辅助的信号反应。当目标 p53 基因存在时,p53 与 L1 和 L2 链之间的相互作用会启动催化发夹组装,并随后暴露 P3 探针中的 DNA 酶。暴露的 DNA 酶与 P4 探针的环状区域结合,产生一个挑刺位点,从而释放出大量与 P4 茎部结合的 ATMND。这导致荧光反应放大,成为检测 p53 基因的荧光信号。这种方法可以准确灵敏地鉴定 p53 基因,其线性反应范围为 1 fM 至 1 nM,检测限低至 0.23 fM。此外,这种荧光方法还可用于临床样本的检测,并具有良好的回收率。最重要的是,这种多功能平台可以通过改变相应的识别单元来分析不同的靶标,在肿瘤发生分析的床旁检测方面显示出巨大的潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
A simple and enzyme-free method for sensitive p53 analysis based on DNAzyme-mediated signal amplification
There is an urgent demand for a simple yet extremely accurate biosensor to analyze tumorigenesis. Herein, we present a novel fluorescent and enzyme-free approach for detecting p53 gene cascading proximity ligation-mediated catalytic hairpin assembly and DNAzyme-assisted signal reaction. When the target p53 gene is present, the interaction between p53 and L1 and L2 chains initiates catalytic hairpin assembly and subsequently exposes DNAzyme in the P3 probe. The exposed DNAzyme binds with the loop region of the P4 probe and generates a nicking site, resulting in the release of a significant amount of ATMND that is conjugated in the stem section of P4. This leads to an amplified fluorescence response, which serves as a fluorescence signal for the detection of the p53 gene. This method allows for the accurate and sensitive identification of the p53 gene, exhibiting a linear reaction range of 1 fM to 1 nM, with a limit of detection as low as 0.23 fM. Furthermore, this fluorescent method has been utilized for the examination of clinical samples with a favorable recovery rate. Crucially, this versatile platform may be expanded to analyze different targets by changing the corresponding recognition unit, showing great potential for point-of-care testing in tumorigenesis analysis.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytical biochemistry
生物-分析化学
CiteScore
5.70
自引率
0.00%
发文量
283
审稿时长
44 days
期刊介绍:
The journal''s title Analytical Biochemistry: Methods in the Biological Sciences declares its broad scope: methods for the basic biological sciences that include biochemistry, molecular genetics, cell biology, proteomics, immunology, bioinformatics and wherever the frontiers of research take the field.
The emphasis is on methods from the strictly analytical to the more preparative that would include novel approaches to protein purification as well as improvements in cell and organ culture. The actual techniques are equally inclusive ranging from aptamers to zymology.
The journal has been particularly active in:
-Analytical techniques for biological molecules-
Aptamer selection and utilization-
Biosensors-
Chromatography-
Cloning, sequencing and mutagenesis-
Electrochemical methods-
Electrophoresis-
Enzyme characterization methods-
Immunological approaches-
Mass spectrometry of proteins and nucleic acids-
Metabolomics-
Nano level techniques-
Optical spectroscopy in all its forms.
The journal is reluctant to include most drug and strictly clinical studies as there are more suitable publication platforms for these types of papers.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


