基于ssDNA杂交的RT-RAA横向流动检测读数用于检测RNA病毒--以SARS-CoV-2为例
IF 8
1区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
摘要
反转录重组酶辅助扩增(RT-RAA)与核酸侧流检测(NALFA)的结合是一种用于SARS-CoV-2等病毒的床旁检测的有效策略。在这项工作中,我们开发了一种结合 RT-RAA 和 NALFA 的终点诊断方法,该方法基于 ssDNA 杂交,完全依靠核酸的相互作用产生特异性信号。此外,还优化了 RT-RAA 扩增条件,包括反应温度和时间。组装了侧流条带并调整了运行条件。选出了尾端引物的最佳组合,所产生的扩增子在试纸条上被成功检测到。所开发的 RT-RAA-NALFA 方法的检测限(LOD)约为 12 个 RNA 拷贝/微升,除其他 SARS-CoV 种类外,检测 16 种不同病原体的 DNA/RNA 时未发现交叉反应,因此具有良好的分析灵敏度和特异性。通过 RT-RAA-NALFA 对 50 份临床样本进行盲测,结果显示与实时 RT-PCR 的结果高度一致(18/21 份阳性样本和 28/29 份阴性样本被正确识别)。这些结果使总体临床灵敏度达到约 86%,总体临床特异性达到约 97%。在实时 RT-PCR 中,当病毒载量对应的定量周期(Cq)低于 30 时,临床灵敏度值增加到约 94%。这说明该检测方法适用于有效检测 SARS-CoV-2 等 RNA 病毒。本文章由计算机程序翻译,如有差异,请以英文原文为准。
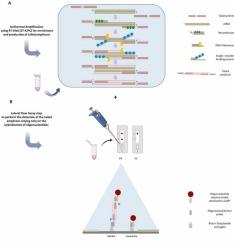
RT-RAA with a lateral flow assay readout based on ssDNA hybridization for detection of RNA viruses – the case of SARS-CoV-2
The conjugation of Reverse-Transcription Recombinase-Aided Amplification (RT-RAA) and Nucleic Acid Lateral Flow Assays (NALFAs) is a useful strategy for the point-of-care detection of viruses like SARS-CoV-2. In this work we developed an end-point diagnostic method combining RT-RAA and NALFA that is based on the hybridization of ssDNA and relies exclusively on the interaction of nucleic acids for producing a specific signal. Different tails were used to modify a previously published set of primers targeting the RdRp gene, enabling the test of distinct sequences and lengths in terms of amplification efficiency. Additionally, RT-RAA amplification conditions, including the temperature and time of reaction, were optimized. Lateral flow strips were assembled and running conditions were adjusted. The best combination of tailed primers was selected and the resulting amplicons were successfully detected in the test strip. The developed RT-RAA-NALFA method showed a limit of detection (LOD) of ∼12 RNA copies/µL and no cross-reactivity was seen testing the DNA/RNA of 16 different pathogens except for other SARS-CoV species, thus exhibiting good analytical sensitivity and specificity. The blind test of 50 clinical samples by RT-RAA-NALFA, showed a high agreement (18/21 positives and 28/29 negatives were correctly identified) with real-time RT-PCR results. These results lead to an overall clinical sensitivity of ∼86 % and an overall clinical specificity of ∼97 %. The clinical sensitivity value increases to ∼94 % for viral loads corresponding to a quantification cycle (Cq) below 30 in real-time RT-PCR. This emphasizes the suitability of the test for the efficient detection of RNA viruses like SARS-CoV-2.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Sensors and Actuators B: Chemical
工程技术-电化学
CiteScore
14.60
自引率
11.90%
发文量
1776
审稿时长
3.2 months
期刊介绍:
Sensors & Actuators, B: Chemical is an international journal focused on the research and development of chemical transducers. It covers chemical sensors and biosensors, chemical actuators, and analytical microsystems. The journal is interdisciplinary, aiming to publish original works showcasing substantial advancements beyond the current state of the art in these fields, with practical applicability to solving meaningful analytical problems. Review articles are accepted by invitation from an Editor of the journal.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


