烟草的全基因组组装揭示了中心粒的遗传和表观遗传景观
IF 15.8
1区 生物学
Q1 PLANT SCIENCES
引用次数: 0
摘要
烟草属(Nicotiana benthamiana)是植物生物学中广泛采用的模式生物。尽管最近对其进行了一些改进,但其完整的基因组组装仍然不可用。为了进一步提高其实用性,我们生成了全四倍体 N. benthamiana 的 2.85 Gb 基因组,并对其进行了分期。我们发现,尽管茄科植物的中心粒普遍由 Ty3/Gypsy 逆转录子主导,但在 N. benthamiana 中,基于卫星的中心粒却出人意料地普遍,19 个中心粒中有 11 个具有兆级规模的卫星阵列。有趣的是,富含卫星和不含卫星的中心粒被不同的吉普赛逆转座子广泛侵染,而 CENH3 蛋白更倾向于占据这些中心粒,这表明它们在中心粒功能中起着关键作用。我们证明核糖体 DNA 是中心粒卫星的主要来源,线粒体 DNA 可作为中心粒的核心成分。亚基因组分析表明,卫星阵列的出现可能推动了新中心粒的形成。总之,我们认为N. benthamiana的中心粒是通过新中心粒形成、卫星扩展、反转座子富集和mtDNA整合进化而来的。本文章由计算机程序翻译,如有差异,请以英文原文为准。
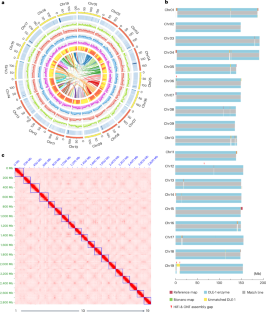

The complete genome assembly of Nicotiana benthamiana reveals the genetic and epigenetic landscape of centromeres
Nicotiana benthamiana is a model organism widely adopted in plant biology. Its complete assembly remains unavailable despite several recent improvements. To further improve its usefulness, we generate and phase the complete 2.85 Gb genome assembly of allotetraploid N. benthamiana. We find that although Solanaceae centromeres are widely dominated by Ty3/Gypsy retrotransposons, satellite-based centromeres are surprisingly common in N. benthamiana, with 11 of 19 centromeres featured by megabase-scale satellite arrays. Interestingly, the satellite-enriched and satellite-free centromeres are extensively invaded by distinct Gypsy retrotransposons which CENH3 protein more preferentially occupies, suggestive of their crucial roles in centromere function. We demonstrate that ribosomal DNA is a major origin of centromeric satellites, and mitochondrial DNA could be employed as a core component of the centromere. Subgenome analysis indicates that the emergence of satellite arrays probably drives new centromere formation. Altogether, we propose that N. benthamiana centromeres evolved via neocentromere formation, satellite expansion, retrotransposon enrichment and mtDNA integration. This study generates and phases the complete genome assembly of a model plant Nicotiana benthamiana, revealing insights into the structure, epigenetic landscape and evolutionary dynamics of its centromeres following allotetraploidization.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


