DigFrag 作为一种数字碎裂方法,用于基于人工智能的药物设计
IF 5.9
2区 化学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
摘要
片段药物设计(FBDD)在药物发现和开发领域发挥着举足轻重的作用。构建高质量的片段库是 FBDD 的关键步骤。传统的片段分析方法往往依赖于僵化的规则和化学直觉,限制了其对不同分子结构的适应性。人工智能(AI)技术的快速发展为重新思考传统方法提供了一个变革性的机会。在这里,我们介绍一种数字片段分析方法 DigFrag,它通过聚焦分子图中的局部结构来突出重要的子结构。此外,我们还将机器智能和人类专业知识分割的片段输入深度生成模型,以比较不同来源数据的偏好。实验结果表明,由 DigFrag 分割的片段结构多样性更高,基于这些片段生成的化合物更理想。这些结果还表明,基于人工智能方法生成的数据可能更适合人工智能模型。此外,基于各种片段技术开发了一个名为 MolFrag ( https://dpai.ccnu.edu.cn/MolFrag/ ) 的用户友好型平台,以支持分子片段分析。基于片段的药物设计在药物发现和开发领域发挥着举足轻重的作用,然而,构建高质量的片段库是一个关键但又极具挑战性的步骤。在此,作者开发了一种基于图注意机制的数字片段分析方法 DigFrag,该方法显示了片段结构的更高多样性和基于人工智能的药物设计的更高适用性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
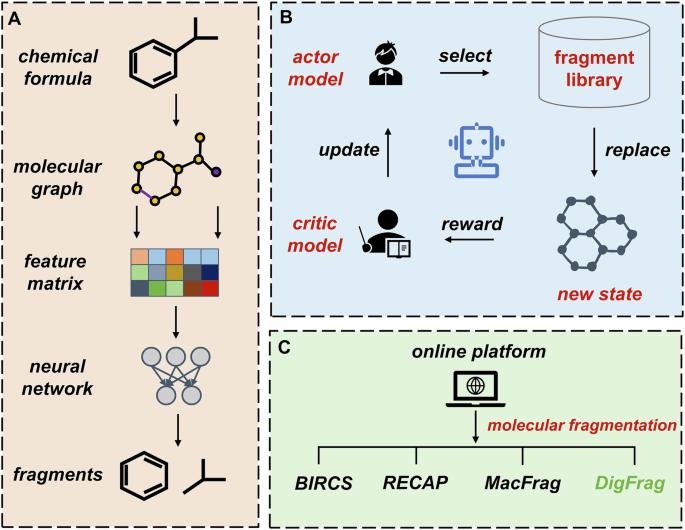
DigFrag as a digital fragmentation method used for artificial intelligence-based drug design
Fragment-Based Drug Design (FBDD) plays a pivotal role in the field of drug discovery and development. The construction of high-quality fragment libraries is a critical step in FBDD. Conventional fragmentation approaches often rely on rigid rules and chemical intuition, limiting their adaptability to diverse molecular structures. The rapid development of Artificial Intelligence (AI) technology offers a transformative opportunity to rethink traditional methods. Here, we present DigFrag, a digital fragmentation method that highlights important substructures by focusing locally within the molecular graph. In addition, we feed the fragments segmented by machine intelligence and human expertise into the deep generative model to compare the preference for data from different sources. Experimental results show that the structural diversity of fragments segmented by DigFrag is higher, and more desirable compounds are generated based on these fragments. These results also demonstrate that data generated based on AI methods may be more suitable for AI models. Moreover, a user-friendly platform called MolFrag ( https://dpai.ccnu.edu.cn/MolFrag/ ) is developed based on various fragmentation techniques to support molecular segmentation. Fragment-based drug design plays a pivotal role in the field of drug discovery and development, however, the construction of high-quality fragment libraries is a critical but challenging step. Here, the authors develop DigFrag, a digital fragmentation method based on the graph attention mechanism, showing higher structural diversity of the fragments and higher applicability to artificial intelligence-based drug design.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Communications Chemistry
Chemistry-General Chemistry
CiteScore
7.70
自引率
1.70%
发文量
146
审稿时长
13 weeks
期刊介绍:
Communications Chemistry is an open access journal from Nature Research publishing high-quality research, reviews and commentary in all areas of the chemical sciences. Research papers published by the journal represent significant advances bringing new chemical insight to a specialized area of research. We also aim to provide a community forum for issues of importance to all chemists, regardless of sub-discipline.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


