多组学孟德尔随机化研究确定了酒精使用障碍和问题饮酒的新治疗靶点
IF 21.4
1区 心理学
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
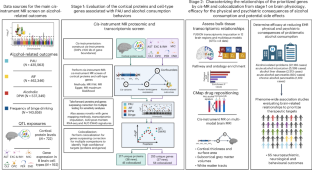
将蛋白质组和转录组数据与问题酒精使用和酒精消费行为的基因结构相结合,可以加深我们的理解,并有助于确定治疗靶点。我们利用基因组学关联研究数据对约 3,500 个皮质蛋白质(N = 722)和 8 种典型脑细胞类型(N = 192)中的约 6,100 个基因与 4 种酒精相关结果(N ≤ 537,349)进行了系统筛选,确定了与这些行为相关的 217 个皮质蛋白质和 255 个细胞类型基因,其中 36 个蛋白质和 37 个细胞类型基因是新发现的。虽然蛋白质组和转录组目标之间的重叠有限,但下游神经影像学发现了共同的神经生理通路。与独立基因组关联研究数据的共定位进一步确定了 16 个蛋白质(包括 CAB39L 和 NRBP1)和 12 个细胞类型基因的优先级,这与 mTOR 信号传导等机制有关。此外,还发现 SAMHD1、VIPAS39、NUP160 和 INO80E 等基因具有良好的神经精神特征。这些研究结果提供了有关问题酒精使用和酒精消费行为的基因图谱,为未来的研究提供了有前景的治疗目标。本文章由计算机程序翻译,如有差异,请以英文原文为准。

A multi-omics Mendelian randomization study identifies new therapeutic targets for alcohol use disorder and problem drinking
Integrating proteomic and transcriptomic data with genetic architectures of problematic alcohol use and alcohol consumption behaviours can advance our understanding and help identify therapeutic targets. We conducted systematic screens using genome-wise association study data from ~3,500 cortical proteins (N = 722) and ~6,100 genes in 8 canonical brain cell types (N = 192) with 4 alcohol-related outcomes (N ≤ 537,349), identifying 217 cortical proteins and 255 cell-type genes associated with these behaviours, with 36 proteins and 37 cell-type genes being new. Although there was limited overlap between proteome and transcriptome targets, downstream neuroimaging revealed shared neurophysiological pathways. Colocalization with independent genome-wise association study data further prioritized 16 proteins, including CAB39L and NRBP1, and 12 cell-type genes, implicating mechanisms such as mTOR signalling. In addition, genes such as SAMHD1, VIPAS39, NUP160 and INO80E were identified as having favourable neuropsychiatric profiles. These findings provide insights into the genetic landscapes governing problematic alcohol use and alcohol consumption behaviours, highlighting promising therapeutic targets for future research. This study examines links between genetics and alcohol use, and identifies 217 proteins in the human cortex and 255 genes at single-cell resolution. Leveraging population genetics with proteomic and transcriptomic data opens new paths in addiction science.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Human Behaviour
Psychology-Social Psychology
CiteScore
36.80
自引率
1.00%
发文量
227
期刊介绍:
Nature Human Behaviour is a journal that focuses on publishing research of outstanding significance into any aspect of human behavior.The research can cover various areas such as psychological, biological, and social bases of human behavior.It also includes the study of origins, development, and disorders related to human behavior.The primary aim of the journal is to increase the visibility of research in the field and enhance its societal reach and impact.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


