关键的汞甲基化活性微生物及其对稻田土壤中甲基汞生成的协同效应
IF 12.2
1区 环境科学与生态学
Q1 ENGINEERING, ENVIRONMENTAL
引用次数: 0
摘要
稻田土壤中含有神经毒性甲基汞(MeHg)的水稻污染问题日益受到全球关注。确定这些土壤中负责汞(Hg)甲基化的微生物,对于控制食物链中的汞污染和减轻对健康的影响至关重要。目前的研究往往侧重于汞甲基化微生物的总量,而忽视了活性微生物的贡献,这可能导致高估或忽视微生物在水稻田土壤汞甲基化中的具体作用。本研究结合 DNA-SIP、汞同位素标记和汞甲基化基因测序技术,对水稻田土壤中活跃的汞甲基化微生物进行了鉴定。我们的研究结果表明,Geobacter 和 Anaerolinea 是水稻田土壤污染梯度中关键的活性汞甲基化微生物。对中国主要水稻生产省份土壤的转录组分析证实了这些微生物的广泛参与和协同作用。微生物培养进一步验证了它们之间的相互作用能显著提高汞的甲基化,Me198Hg 浓度比单独使用 Geobacter 时提高了 2.8 倍,比单独使用 Anaerolinea 时提高了 5.2 倍。这些发现加深了我们对稻田土壤中微生物汞甲基化作用的了解,为准确预测土壤甲基汞负荷、稻谷甲基汞污染和人类接触甲基汞的风险提供了重要依据。本文章由计算机程序翻译,如有差异,请以英文原文为准。
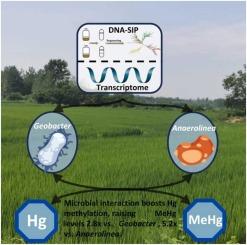
Key active mercury methylating microorganisms and their synergistic effects on methylmercury production in paddy soils
Rice contamination with neurotoxic methylmercury (MeHg) from paddy soils is an escalating global concern. Identifying the microorganisms responsible for mercury (Hg) methylation in these soils is essential for controlling Hg contamination in the food chain and mitigating health impacts. Current research often focuses on total Hg-methylating microorganisms, overlooking the contributions of active ones, which can lead to either overestimating or neglecting the specific roles of microorganisms in Hg methylation within paddy soils. In this study, active Hg-methylating microorganisms in paddy soils were identified using a combination of DNA-SIP, Hg isotope labelling, and Hg methylation gene sequencing techniques. Our findings revealed that Geobacter and Anaerolinea are pivotal active Hg-methylating microorganisms across a contamination gradient in paddy soils. Transcriptomic analysis of soils from major rice-producing provinces in China confirmed the widespread and synergistic involvement of these microorganisms. Microbial incubation further validated their interaction significantly enhances Hg methylation, with Me198Hg concentrations increasing 2.8-fold compared to Geobacter alone and 5.2-fold compared to Anaerolinea alone. These findings enhance our understanding of microbial Hg methylation in paddy soils, providing critical insights for accurately predicting soil MeHg load, rice grain MeHg contamination, and human MeHg exposure risks.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Hazardous Materials
工程技术-工程:环境
CiteScore
25.40
自引率
5.90%
发文量
3059
审稿时长
58 days
期刊介绍:
The Journal of Hazardous Materials serves as a global platform for promoting cutting-edge research in the field of Environmental Science and Engineering. Our publication features a wide range of articles, including full-length research papers, review articles, and perspectives, with the aim of enhancing our understanding of the dangers and risks associated with various materials concerning public health and the environment. It is important to note that the term "environmental contaminants" refers specifically to substances that pose hazardous effects through contamination, while excluding those that do not have such impacts on the environment or human health. Moreover, we emphasize the distinction between wastes and hazardous materials in order to provide further clarity on the scope of the journal. We have a keen interest in exploring specific compounds and microbial agents that have adverse effects on the environment.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


