赖氨酸脱乙酰酶 sirtuin 活性的生物发光测定
IF 7.2
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
赖氨酸酰化可指导蛋白质的功能、定位和相互作用。Sirtuins 对赖氨酸进行脱乙酰化,以维持细胞的平衡,而它们的异常表达则是包括癌症在内的多种疾病的发病机理之一。测量 Sirtuins 的活性对于探索其作为治疗靶点的潜力至关重要,但精确量化却很有挑战性。我们开发了 "SIRTify",这是一种用于测量体内外 sirtuin 活性的高灵敏度检测方法。SIRTify 基于 NanoLuc 荧光素酶的分裂版本,由一个截短的、无催化活性的 N 端分子(LgBiT)与一个高亲和力的 C 端肽(p86)互补形成活性荧光素酶。p86 中两个赖氨酸的酰化会破坏与 LgBiT 的结合并减弱荧光。sirtuin 的脱酰基作用可重建 p86 并恢复结合,从而产生与 sirtuin 活性成比例的发光信号。测量结果准确反映了所报道的 sirtuin 对赖氨酸酰化的特异性,并证实了 sirtuin 调节剂的作用。SIRTify 可量化赖氨酸脱酰化动态,并可用于监测其他翻译后修饰。本文章由计算机程序翻译,如有差异,请以英文原文为准。

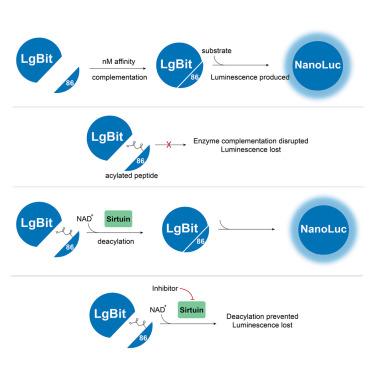
Bioluminescence assay of lysine deacylase sirtuin activity
Lysine acylation can direct protein function, localization, and interactions. Sirtuins deacylate lysine toward maintaining cellular homeostasis, and their aberrant expression contributes to the pathogenesis of multiple conditions, including cancer. Measuring sirtuins’ activity is essential to exploring their potential as therapeutic targets, but accurate quantification is challenging. We developed “SIRTify”, a high-sensitivity assay for measuring sirtuin activity in vitro and in vivo. SIRTify is based on a split-version of the NanoLuc luciferase consisting of a truncated, catalytically inactive N-terminal moiety (LgBiT) that complements with a high-affinity C-terminal peptide (p86) to form active luciferase. Acylation of two lysines within p86 disrupts binding to LgBiT and abates luminescence. Deacylation by sirtuins reestablishes p86 and restores binding, generating a luminescence signal proportional to sirtuin activity. Measurements accurately reflect reported sirtuin specificity for lysine-acylations and confirm the effects of sirtuin modulators. SIRTify quantifies lysine deacylation dynamics and may be adaptable to monitoring additional post-translational modifications.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell Chemical Biology
Biochemistry, Genetics and Molecular Biology-Molecular Medicine
CiteScore
14.70
自引率
2.30%
发文量
143
期刊介绍:
Cell Chemical Biology, a Cell Press journal established in 1994 as Chemistry & Biology, focuses on publishing crucial advances in chemical biology research with broad appeal to our diverse community, spanning basic scientists to clinicians. Pioneering investigations at the chemistry-biology interface, the journal fosters collaboration between these disciplines. We encourage submissions providing significant conceptual advancements of broad interest across chemical, biological, clinical, and related fields. Particularly sought are articles utilizing chemical tools to perturb, visualize, and measure biological systems, offering unique insights into molecular mechanisms, disease biology, and therapeutics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


