大规模外显子组测序在 394 005 名英国人中发现了 18 个神经质新基因
IF 15.9
1区 心理学
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
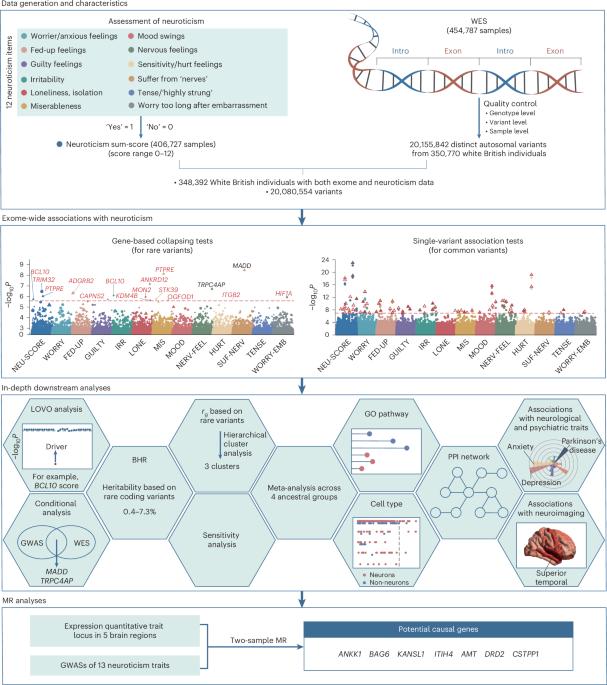
现有的神经质基因研究主要局限于常见变异。在这里,我们对英国生物库中的英国白人进行了大规模外显子组分析,揭示了编码变异在神经质中的作用。对于罕见变异,折叠分析发现了 14 个与神经质相关的基因。其中,12个基因(PTPRE、BCL10、TRIM32、ANKRD12、ADGRB2、MON2、HIF1A、ITGB2、STK39、CAPNS2、OGFOD1和KDM4B)是新基因,其余的基因(MADD和TRPC4AP)与常见变异有趋同证据。据估计,神经质罕见编码变异的遗传率高达 7.3%。对于常见变异,我们确定了 78 个显著关联,其中涉及 6 个未报告的基因。随后,我们对英国生物库中的其他四个血统和 23andMe 样本的汇总数据进行了荟萃分析,复制了这些变异。此外,这些变异对神经精神疾病、认知能力和大脑结构有着广泛的影响。我们的发现加深了人们对神经质遗传结构的理解,并为未来的机理研究提供了潜在的目标。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Large-scale exome sequencing identified 18 novel genes for neuroticism in 394,005 UK-based individuals
Existing genetic studies of neuroticism have been largely limited to common variants. Here we performed a large-scale exome analysis of white British individuals from UK Biobank, revealing the role of coding variants in neuroticism. For rare variants, collapsing analysis uncovered 14 neuroticism-associated genes. Among these, 12 (PTPRE, BCL10, TRIM32, ANKRD12, ADGRB2, MON2, HIF1A, ITGB2, STK39, CAPNS2, OGFOD1 and KDM4B) were novel, and the remaining (MADD and TRPC4AP) showed convergent evidence with common variants. Heritability of rare coding variants was estimated to be up to 7.3% for neuroticism. For common variants, we identified 78 significant associations, implicating 6 unreported genes. We subsequently replicated these variants using meta-analysis across other four ancestries from UK Biobank and summary data from 23andMe sample. Furthermore, these variants had widespread impacts on neuropsychiatric disorders, cognitive abilities and brain structure. Our findings deepen the understanding of neuroticism’s genetic architecture and provide potential targets for future mechanistic research. In 394,005 UK individuals with exome sequencing data, Wu et al. find 56 neuroticism-associated genes (18 novel) and quantify the heritability explained by rare coding variants, highlighting the association of coding variants with neuroticism.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Human Behaviour
Psychology-Social Psychology
CiteScore
36.80
自引率
1.00%
发文量
227
期刊介绍:
Nature Human Behaviour is a journal that focuses on publishing research of outstanding significance into any aspect of human behavior.The research can cover various areas such as psychological, biological, and social bases of human behavior.It also includes the study of origins, development, and disorders related to human behavior.The primary aim of the journal is to increase the visibility of research in the field and enhance its societal reach and impact.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


