依赖自噬的剪接控制引导衰老过程中的炎症翻译
IF 10.7
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
摘要
细胞蛋白质组决定着细胞的功能状态,并常常直接影响病理状态。自噬主要通过溶酶体降解受损或不必要的蛋白质来塑造细胞蛋白质组。在这里,我们展示了自噬通过将选择性蛋白质降解与替代剪接结合起来,引导衰老特异性转译组助长炎症。在衰老过程中,RNA剪接发生了显著变化,其中一些竟然依赖于自噬,包括翻译调节因子EIF4H的第5外显子跳过。系统翻译组剖析表明,这一事件是衰老期翻译偏向炎症的关键。自噬通过自噬受体 NBR1 选择性地降解剪接调节因子富脯氨酸和谷氨酰胺(SFPQ),从而促进了这些变化。这些以自噬为中心的炎症控制似乎在人体组织衰老和癌症过程中是一致的。我们的研究强调了自噬在细胞蛋白质组按需功能重塑中的作用,以及自噬、替代剪接和炎症翻译之间的相互影响。本文章由计算机程序翻译,如有差异,请以英文原文为准。
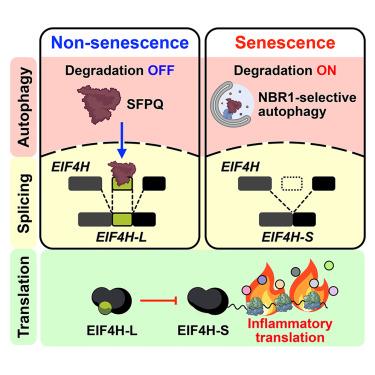
Autophagy-dependent splicing control directs translation toward inflammation during senescence
The cellular proteome determines the functional state of cells and is often skewed to direct pathological conditions. Autophagy shapes cellular proteomes primarily through lysosomal degradation of either damaged or unnecessary proteins. Here, we show that autophagy directs the senescence-specific translatome to fuel inflammation by coupling selective protein degradation with alternative splicing. RNA splicing is significantly altered during senescence, some of which surprisingly depend on autophagy, including exon 5 skipping of the translation regulator EIF4H. Systematic translatome profiling indicates that this event is key to the translational bias toward inflammation in senescence. Autophagy promotes these changes by selectively degrading the splicing regulator splicing factor proline and glutamine rich (SFPQ) via the autophagy receptor NBR1. These autophagy-centric inflammatory controls appear to be conserved during human tissue aging and cancer. Our work highlights the role of autophagy in the on-demand functional remodeling of cellular proteomes as well as the crosstalk between autophagy, alternative splicing, and inflammatory translation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Developmental cell
生物-发育生物学
CiteScore
18.90
自引率
1.70%
发文量
203
审稿时长
3-6 weeks
期刊介绍:
Developmental Cell, established in 2001, is a comprehensive journal that explores a wide range of topics in cell and developmental biology. Our publication encompasses work across various disciplines within biology, with a particular emphasis on investigating the intersections between cell biology, developmental biology, and other related fields. Our primary objective is to present research conducted through a cell biological perspective, addressing the essential mechanisms governing cell function, cellular interactions, and responses to the environment. Moreover, we focus on understanding the collective behavior of cells, culminating in the formation of tissues, organs, and whole organisms, while also investigating the consequences of any malfunctions in these intricate processes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


