古生物 NusA2 是真核生物核糖体蛋白 eS7 的祖先
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
N-utilization substance A(NusA)是一种在细菌基因表达中具有多种功能的调控因子。古细菌编码两种保守的小蛋白 NusA1 和 NusA2,其结构域与 NusA 的两个 RNA 结合 K 同源(KH)结构域同源。在此,我们报告了酸性硫球菌(Sulfolobus acidocaldarius)和溶藻糖球菌(Saccharolobus solfataricus)的 NusA2 晶体结构,它们的结构分别为 3.1 Å 和 1.68 Å。NusA2 由一个 N 端锌指和两个缺乏 GXXG 标志的 KH 样结构域组成。尽管失去了 GXXG 基序,NusA2 仍能结合单链 RNA。锌指结构域的突变损害了 NusA2 在高温下的结构完整性,分子动力学模拟表明,锌结合提供了一个能量屏障,阻止该结构域达到展开状态。对 KH 样结构域的结构指导系统发育分析支持 NusA2 支系是真核生物中核糖体蛋白 eS7 的祖先这一观点,这意味着 NusA2 在翻译中的潜在作用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
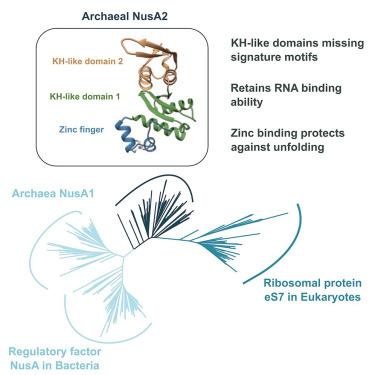
Archaeal NusA2 is the ancestor of ribosomal protein eS7 in eukaryotes
N-utilization substance A (NusA) is a regulatory factor with pleiotropic functions in gene expression in bacteria. Archaea encode two conserved small proteins, NusA1 and NusA2, with domains orthologous to the two RNA binding K Homology (KH) domains of NusA. Here, we report the crystal structures of NusA2 from Sulfolobus acidocaldarius and Saccharolobus solfataricus obtained at 3.1 Å and 1.68 Å, respectively. NusA2 comprises an N-terminal zinc finger followed by two KH-like domains lacking the GXXG signature. Despite the loss of the GXXG motif, NusA2 binds single-stranded RNA. Mutations in the zinc finger domain compromise the structural integrity of NusA2 at high temperatures and molecular dynamics simulations indicate that zinc binding provides an energy barrier preventing the domain from reaching unfolded states. A structure-guided phylogenetic analysis of the KH-like domains supports the notion that the NusA2 clade is ancestral to the ribosomal protein eS7 in eukaryotes, implying a potential role of NusA2 in translation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


